ABOUT

The Department of Biochemistry and Molecular Biology has existed since 2012, when the Department of Biochemistry and Biophysics (founded in 1962) of the former Faculty of Natural Sciences of Vilnius University was divided into two departments – the Department of Biochemistry and Molecular Biology and the Department of Neurobiology and Biophysics.
We are proud of the hundreds of graduates from the Department – biochemists and molecular biologists – who work at Lithuanian and foreign universities, research centers, business enterprises, and state institutions, making Lithuania well-known in the fields of Life sciences, technology, and innovation.
We organize and participate in biochemistry studies at the I-III levels and molecular biology studies at the I-II levels at Vilnius University. The Department's scientists conduct research on various topics in the fields of biochemistry, molecular biology, and cell biology.
RESEARCH
Molecular Mechanisms of Bacterial Antibiotic Resistance and Pathogenesis
Principal investigators
Prof. Edita Sužiedėlienė
Prof. Julija Armalytė
Dr. Jūratė Skerniškytė
In collaboration (VU, Faculty of Physics): Dr. Irina Buchovec
Group members
Dr. Danutė Labeikytė
Assoc. Prof. Arvydas Markuckas
Prof. Kęstutis Sužiedėlis
Ramutė Pagalytė
PhD students
Laurita Klimkaitė
Tomas Liveikis
Ignas Ragaišis
Research topics
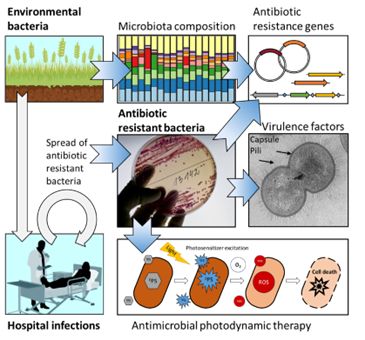
Our research focuses on understanding the molecular basis of bacterial antibiotic resistance in both clinical and environmental settings, with an emphasis on novel resistance mechanisms and bacterial cell features that contribute to pathogenesis.
Genetic elements responsible for the spread and evolution of antimicrobial resistance in the clinic and the environment
Infections caused by a group of gram-negative bacteria resistant to nearly all currently available antibiotics are a serious concern in clinical settings worldwide. Bacteria, previously considered non-pathogenic due to their ability to acquire multidrug resistance and virulence traits, are becoming one of the most essential hospital infection agents. The opportunistic pathogen Acinetobacter baumannii causes a variety of difficult-to-treat nosocomial infections in critically ill patients. The characteristic features of A. baumannii include the ability to withstand prolonged periods of dryness, form biofilms on various surfaces, including medical equipment, upregulate intrinsic resistance mechanisms, and acquire new resistance genes through plasmids, as well as the ability to adhere to and colonize host cells. Another opportunistic pathogen, Stenotrophomonas maltophilia, is receiving increased attention in the clinic due to its innate multidrug resistance, which causes difficult-to-treat infections and a lack of knowledge about the molecular mechanisms underlying its pathogenicity. Understanding the molecular basis of pathogenesis and evaluating the impact of mobile genetic elements on bacterial adaptation may provide novel insights into the pathogenesis process and suggest the development of novel antibacterial strategies.
The interplay between opportunistic bacteria and the innate immunity
The virulence mechanisms employed by opportunistic pathogens enable them to infect the host efficiently. Our research has revealed two important bacterial features in the pathogenesis of opportunistic bacteria: 1) capsular polysaccharides (CPS) and 2) outer membrane vesicles (OMV). We demonstrated that CPS efficiently protects A. baumannii from environmental stress and phagocytosis by immune cells. Additionally, A. baumannii produces OMVs that encapsulate antibiotic-degrading enzymes, making OMV secretion a powerful strategy for inactivating antimicrobials at a distance. Our investigations have revealed that secreted OMV can also modulate the inflammatory response in macrophages. We aim to understand how bacteria modulate the immune response by producing CPS and OMV.
Alternative antibacterial treatment techniques
Antimicrobial photodynamic therapy (aPDT) is a biophotonic technology that can be used to treat multidrug-resistant bacteria as a complementary or alternative to antibiotics. aPDT is based on the interaction of a photosensitizer, molecular oxygen, and low doses of light, generating reactive oxygen species (ROS) that damage bacterial cells. Our recent work demonstrates that aPDT, using photosensitizers such as chlorophyllin (Chl), riboflavin (Rf), and 5-aminolevulinic acid (ALA), can be used against biofilms of A. baumannii and S. maltophilia. A combination of aPDT with antibiotics could enhance antibacterial effect, leading to the complete neutralisation of pathogens and preventing the recurrence of infection.
Microbial diversity and spread of antibiotic resistance in the environment
Microorganisms are ubiquitous and play a leading role in countless natural environmental processes. However, the biodiversity in soil, water, and other bacterial habitats can be threatened by human activities and disruptions to established natural processes. Apart from soil or water body biodiversity management being vital for agricultural purposes, these habitats are also considered favourable environments for the evolution and development of antimicrobial resistance due to their high complexity and ongoing competition among microorganisms. We aim to evaluate variation in microbial composition across different environmental conditions (e.g., intensive agricultural lands, aquaculture, polluted areas), with a focus on the potential spread of clinically significant antibiotic resistance genes (ARGs). The comparison of ARGs' composition, as well as their mobilization potential, could show the impact of anthropogenic activities on ecosystems and the possibility of transferring ARGs to human hosts.
Analysis of the human airway microbiome
The human microbiome, including that of the respiratory tract, is being described and characterized in an increasing number of studies; however, the composition and effects of the healthy or disturbed microbiome on pulmonary health, as well as its interactions with the host, are only beginning to be elucidated. In our studies, we aim to investigate the upper respiratory tract as a less invasive alternative for understanding lung health and identifying potential biomarkers linked to the disease. The microbial diversity of the upper and lower airways is determined using full-length 16S rRNA gene amplicon sequencing with Oxford Nanopore Technologies.
Projects
Researcher Groups Project “Lung microbiome study of Lithuanian tuberculosis patients” funded by The Research Council of Lithuania (S-MIP-24-70), researcher Dr. J. Armalytė, (2024–2027).
Science promotion fund of Vilnius University project “Application of bacterial outer membrane vesicles to deliver antibiotics and photoactive substances for combined therapy against biofilms formed by opportunistic bacteria” (MSF-JM-04/2024), principal investigator Dr. J. Skerniškytė, project participant Dr. I. Buchovec (2024–2025).
Researcher Groups Project “Investigation of capsular polysaccharides and outer membrane vesicles as virulence factors in pathogenesis of opportunistic bacteria” funded by The Research Council of Lithuania (S-MIP-23-109), principal investigator Dr. J. Skerniškytė (2023–2026).
Science promotion fund of Vilnius University project “Prevalence of genetic determinants of antibiotic resistance in opportunistic pathogens Acinetobacter baumannii and Stenotrophomonas maltophilia“ (MSF-JM-12/2023), principal investigator L. Klimkaitė, project participant T. Liveikis (2023–2024).
Science promotion fund of Vilnius University project “Synergistic Antibiotic-Photodynamic Therapy Combination in Inactivation of Opportunistic Pathogen Stenotrophomonas maltophilia”, (MSF-JM-3/2021), principal investigator Dr. I. Buchovec, project participant L. Klimkaitė (2021–2022).
Programme for the European Union Funds Investments in Lithuania, Funding instrument - European Regional Development Fund, measure 01.2.2-LMT-K-718 „Targeted Research in Smart Specialisation Areas“, project „Analysis of Individualized Upper Respiratory Tract Microbiome – a novel tool for diagnostics and healthcare (YourAirwayMicrobiome)“, (01.2.2-LMT-K-718-03-0079), project leader Innovative Medicine Center, project coordinator for Vilnius University Dr. J. Armalytė (2020–2023).
National research program “Healthy ageing”, project “Development of virus-like particles-based vaccine against Acinetobacter baumannii“ (S-SEN20-1), principal investigator Dr. J. Armalytė (2020–2021).
National research program “Sustainability of agro, forest and water ecosystems”, project “The influence of intensive fish farming on aquatic microbiome and resistome“, (S-SIT-20-6), researcher Dr. J. Armalytė (2020–2021).
Laboratory of Nucleic Acids Biochemistry
Principal investigator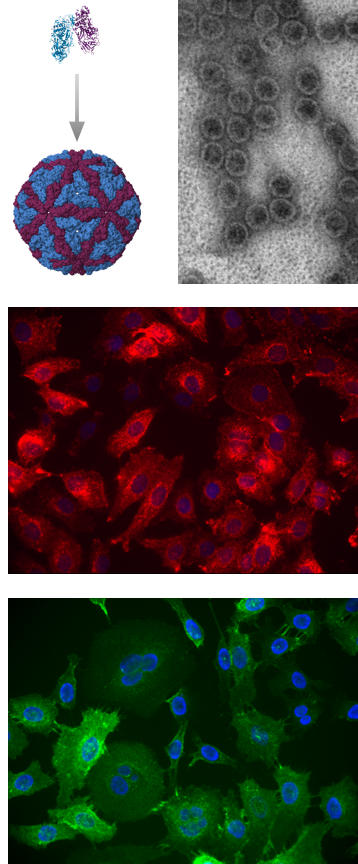
Prof. Saulius Serva
Group members
Dr. Aleksandras Konovalovas
Dr. Algirdas Mikalkėnas
Dr. Enrika Celitan
PhD students
Gerda Skinderytė
Mehvish Mumtaz
Research
The group is actively engaged in research focused on two main directions:
- Molecular mechanisms of innate yeast Saccharomyces viruses
The dsRNA viruses are ubiquitous yet poorly understood benign inhabitants of the yeast. Understanding the relationships between viruses and their hosts, both intra- and extracellular, is crucial for elucidating viral evolutionary pathways and revealing their roles in ecosystems.
To this end, the diversity and functions of fungal dsRNA viruses are addressed, starting with their identification in natural hosts and proceeding to in-depth investigation in our lab.
Our team employs systems biology approaches to investigate the interactions between yeast double-stranded RNA viruses and the host cell. The developed techniques enable genomics, transcriptomics, proteomics, phenomics, and structural analysis aimed at understanding the molecular mechanisms underlying the establishment of yeast viruses. Translational applications of dsRNA virus capsids for facilitated drug delivery
In addition to a basic fundamental focus, we develop the translational horizon of yeast viruses. The engineering, preparation, and application of virus capsids for facilitated drug delivery are actively pursued goals in the formulation of advanced biomedical applications.
Methods
A wide range of methods, from classic to those recently developed, is employed in the lab. We specialize in microbiology, gene and genome engineering, next-generation DNA and RNA sequencing (utilizing Illumina and Oxford Nanopore technologies), bioinformatics, enzymology, advanced microscopy and structural analysis, cell culture applications, and other relevant techniques.
Projects (since 2020)
Research Council of Lithuania: “Microcosmos of flower and honey bee drones and sustainable control of hive health (MICROCOSMOS)”, No. S-MIP-24-55, 2024–2027.
Research Council of Lithuania: “Lung microbiome study of Lithuanian tuberculosis patients”, No. S-MIP-24-70, 2024–2027.
University Excellence Initiative Programme: “Data Center for Machine Learning and Quantum Computing in Natural and Biomedical Sciences”, No.: S-A-UEI-23-11, 2023–2027.
Research Council of Lithuania: “Facilitated delivery of bioactive compounds: versatile toolkit for biomedical applications (DECORATION)”, No. S-MIP-23-28, 2023–2026.
Research Council of Lithuania: “Linkage of biocidal phenomenon of natural yeast and native antiviral systems”, No. S-ST-23-92, 2023–2024.
Research Council of Lithuania: “Biocontrol of wild berry microbiota composition by yeast counterpart (CONFIDENT)”, P-PD-22-058, 2022–2024.
Vilnius University Science promotion programme: “DNA Engineering Tools for Pathogen Identification in Bioarchaeological Samples”, MSF-JM-14/2022, 2022–2023.
VU LSC-EMBL Partnership Institute Associated Research Fellowships: “CRISPR-Based Enrichment Strategy for Targeted Long-Read Sequencing”, 2022–2023.
Research Council of Lithuania: “Individualized Nasal Microbiome Test – Novel Tool for Diagnostics and Health Care (YourAirwayMicrobiome)”, 01.2.2-LMT-K-718-03-0079, 2021–2023.
Ministry of Education, Science and Sports: “System for virus spread control and extreme situation management during COVID-19 epidemics”, Nr. S-DNR-20-2 (2021).
Research Council of Lithuania: “Viractome - integrative approach for functional yeast virus analysis”, No. 09.3.3-LMT-K-712-19-0157, 2020–2022.
European Cooperation in Science and Technology (COST) activity CA17103: “Delivery of antisense RNA therapeutics (DARTER)”, P-COST-20-3, 2020–2022.
Research Council of Lithuania: “Chemical Annotation in the Crystallography Open Database (COD)”, S-MIP-20-21, 2020–2022.
Vilnius University Science Advancement Fund: „Investigation of Totiviridae family virus ScV-LA biogenesis in native environment“, Nr. MSF-JM-7, 2019–2020.
EU Horizon 2020 Research and Innovation Program: “Sonic drilling coupled with automated mineralogy and chemistry On-Line-On-Mine-Real-Time (SOLSA)”, 2016–2020.
Molecular mechanisms of resistance to anticancer treatment
Principal investigator
Dr. Aušra Sasnauskienė
Group members
Dr. Violeta Jonušienė
Dr. Daiva Dabkevičienė
Vilmantė Žitkutė
Eglė Žalytė
Research topics
Research interests of our group:
• Investigation of molecular mechanisms of resistance to anticancer treatment;
• Functional studies of human primary cells.
Investigation of molecular mechanisms of resistance to anticancer treatment
Acquired drug resistance is a major limitation of cancer treatment. Resistance of cancer cells can emerge due to various factors, including alterations in drug transport and metabolism, modification of drug targets, activation of DNA repair, or changes in cell death induction. A deeper understanding of chemoresistant cell physiology, particularly cell death and survival signalling, suggests new potential targets to overcome cancer cell resistance.
We study molecular mechanisms that determine cancer cell susceptibility to agents of chemotherapy, targeted therapy, or immune checkpoint inhibition. Our aim is to identify potential targets for anti-cancer therapy in colorectal and endometrial cancer cells.
In collaboration with researchers at the Institute of Biochemistry, Vilnius University Life Sciences Center, we investigate the use of bacteriophage nanotubes as potential drug carriers. We evaluate the mechanisms of nanotube entry and transport in cancer cells.
Functional analysis of human primary cells
In collaboration with the Institute of Biomedical Sciences, Faculty of Medicine, Vilnius University, we analyze primary cells of patients with rare genetic diseases. We aim to characterize functional changes of primary cells and correlate them with alterations in the genome and transcriptome.
Methods
Culturing of cell cultures and primary cells in 2D and 3D systems; immunoenzymatic assays: Western blot, ELISA; quantitative PCR; subcellular localization studies using confocal microscopy; flow cytometry; functional analysis of gene expression using siRNA and CRISPR-Cas techniques.
Projects (since 2017)
Science promotion fund of Vilnius University project “Application of CRISPR-Cas13 technology in studying mechanisms of chemoresistance”, No. MSF-JM-2/2021. Project leader V. Žitkutė (2021-2022).
Research Council of Lithuania National programme “Healthy ageing” project “Self-assembling Phage Proteins for Targeted Nanomedicine”, No. P-SEN-20-4. Participants: A. Sasnauskienė, V. Žitkutė (2020-2021).
Science promotion fund of Vilnius University project “Genome and transcriptome analysis in pathogenesis studies of rare inherited diseases”, No. MSF-JM-2/2020. Participants: A. Sasnauskienė, V. Žitkutė (2020).
Research Council of Lithuania. Project “Redox Chemistry, Biochemistry and Cytotoxicity of Aromatic Nitrocompounds and N-oxides: a New Look”, No. DOTSUT-34/09.33-LMT-K712-01-0058. Participant V. Jonušienė (2018–2021).
Research Council of Lithuania, National programme “Healthy ageing” project “Novel biomarkers for individualized therapy of colon cancer: proteomics, microRNomics and clinics”, No. SEN-17/2015. Participants V. Jonušienė, A. Sasnauskienė (2015-2018).
Molecular mechanisms of intracellular trafficking
Principal investigator
Prof. Vytautė Starkuvienė Erfle
Research topics
Intracellular trafficking distributes newly synthesized and endocytosed material to diverse cellular destinations, thereby ensuring cellular homeostasis. Deregulation of cargo trafficking leads to an ever-increasing list of diseases, such as cancer or cardiovascular diseases. Intracellular trafficking strongly contributes to cell differentiation and aging processes. Trafficking can be divided into several pathways: secretory mechanisms that transport cargo from the endoplasmic reticulum to the Golgi complex, and from there on to the plasma membrane, lysosomes, or the cell outside. Endocytosis is responsible for the cellular entry of various ligands, growth factors, or viruses. Both secretory and endocytic pathways are closely related to degradation, autophagy, cell death and division, transcription, and translation. Our aim is to analyse how the endocytosis machinery responds to and adapts to changes in the cell's environment. The projects are running at Vilnius University and Heidelberg University (Germany).
Methods
To dissect the complexity of trafficking pathways, we use cell biology, molecular genetics, and biochemistry techniques. The methodological focus in the group lies in high-throughput and high-resolution fluorescence microscopy. We work with cancer and healthy primary cells in 2D and 3D environments
and modify gene, transcript, and protein expression and function through gene editing, RNA interference (RNAi), drug, and antibody-mediated approaches
. We also develop techniques to perform these experiments with little side effects and on varying biological scales.
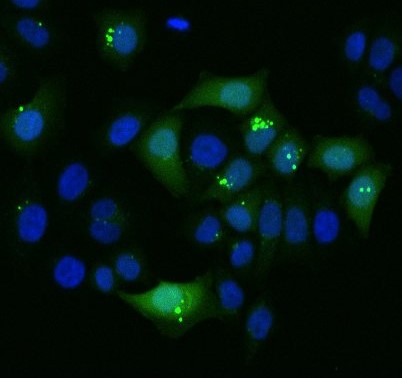
The complexity of endocytosis defines the efficiency of cell modification
Cargo intracellular delivery depends on the effectiveness of endocytosis, which differs in individual cells. GFP protein enters some cells efficiently and localizes to the cytoplasm. In contrast, some cells fail to internalize this cargo. Part of the cells trap GFP in their endosomes after internalization.
Courtesy: Dr S. Liechocki, Heidelberg University
MAIN PUBLICATIONS

Molecular Mechanisms of Bacterial Antibiotic Resistance and Pathogenesis:
Skerniskyte J., Barroso M.V., Chicher J., Hammann P., Demais V., Wright K., Mostowy S., Marteyn B.S. (2025) Neutrophils display antibacterial defense via non-canonical LC3 decoration of extracellular bacteria. Microbes Infect. 105545. 10.1016/j.micinf.2025.105545.
Klimkaitė L., Drevinskaitė R., Krinickis K., Sužiedėlienė E., Armalytė J. (2025) Stenotrophomonas maltophilia of clinical origin display higher temperature tolerance comparing with environmental isolates. Virulence. 16(1):2498669. 10.1080/21505594.2025.2498669.
Konovalovas A., Armalytė J., Klimkaitė L., Liveikis T., Jonaitytė B., Danila E., Bironaitė D., Mieliauskaitė D., Bagdonas E., Aldonytė R. (2024). Insights into respiratory microbiome composition and systemic inflammatory biomarkers of bronchiectasis patients. Microbiol Spectr. 12(12):e0414423. 10.1128/spectrum.04144-23.
Konovalovas A., Armalytė J., Klimkaitė L., Liveikis T., Jonaitytė B., Danila E., Bironaitė D., Mieliauskaitė D., Bagdonas E., Aldonytė R. (2024) Human nasal microbiota shifts in healthy and chronic respiratory disease conditions. BMC Microbiol. 24(1):150. 10.1186/s12866-024-03294-5.
Danne, C., Skerniskyte, J., Marteyn, B., and Sokol, H. (2024). Neutrophils: from IBD to the gut microbiota. Nat Rev Gastroenterol Hepatol 21, 184–197. 10.1038/s41575-023-00871-3.
Armalytė, J., Čepauskas, A., Šakalytė, G., Martinkus, J., Skerniškytė, J., Martens, C., Sužiedėlienė, E., Garcia-Pino, A., and Jurėnas, D. (2023). A polyamine acetyltransferase regulates the motility and biofilm formation of Acinetobacter baumannii. Nat Commun 14, 3531. 10.1038/s41467-023-39316-5.
Klimkaite, L., Liveikis, T., Kaspute, G., Armalyte, J., and Aldonyte, R. (2023). Air pollution-associated shifts in the human airway microbiome and exposure-associated molecular events. Future Microbiol 18, 607–623. 10.2217/fmb-2022-0258.
Klimkaitė, L., Ragaišis, I., Krasauskas, R., Ružauskas, M., Sužiedėlienė, E., and Armalytė, J. (2023). Novel Antibiotic Resistance Genes Identified by Functional Gene Library Screening in Stenotrophomonas maltophilia and Chryseobacterium spp. Bacteria of Soil Origin. Int. J. Mol. Sci. 24, 6037. 10.3390/ijms24076037.
Buchovec, I., Klimkaitė, L., Sužiedėlienė, E., and Bagdonas, S. (2022). Inactivation of Opportunistic Pathogens Acinetobacter baumannii and Stenotrophomonas maltophilia by Antimicrobial Photodynamic Therapy. Microorganisms 10, 506. 10.3390/microorganisms10030506.
Buchovec, I., Vyčaitė, E., Badokas, K., Sužiedelienė, E., and Bagdonas, S. (2022). Application of Antimicrobial Photodynamic Therapy for Inactivation of Acinetobacter baumannii Biofilms. Int. J. Mol. Sci. 24, 722. 10.3390/ijms24010722.
Polmickaitė-Smirnova, E., Buchovec, I., Bagdonas, S., Sužiedėlienė, E., Ramanavičius, A., and Anusevičius, Ž. (2022). Photoinactivation of Salmonella enterica exposed to 5-aminolevulinic acid: Impact of sensitization conditions and irradiation time. Journal of Photochemistry and Photobiology B: Biology 231, 112446. 10.1016/j.jphotobiol.2022.112446.
Lastauskienė, E., Valskys, V., Stankevičiūtė, J., Kalcienė, V., Gėgžna, V., Kavoliūnas, J., Ružauskas, M., and Armalyte, J. (2021). The Impact of Intensive Fish Farming on Microbiome and Resistome of Pond Sediments. Front. Vet. Sci. 8. 10.3389/fvets.2021.673756.
Skerniškytė, J., Karazijaitė, E., Lučiūnaitė, A., and Sužiedėlienė, E. (2021). OmpA Protein-Deficient Acinetobacter baumannii Outer Membrane Vesicles Trigger Reduced Inflammatory Response. Pathogens 10, 407. 10.3390/pathogens10040407.
Klimkaitė, L., Armalytė, J., Skerniškytė, J., and Sužiedėlienė, E. (2020). The Toxin-Antitoxin Systems of the Opportunistic Pathogen Stenotrophomonas maltophilia of Environmental and Clinical Origin. Toxins (Basel) 12. 10.3390/toxins12100635.
Krasauskas, R., Skerniškytė, J., Martinkus, J., Armalytė, J., and Sužiedėlienė, E. (2020). Capsule Protects Acinetobacter baumannii From Inter-Bacterial Competition Mediated by CdiA Toxin. Front Microbiol 11, 1493. 10.3389/fmicb.2020.01493.
Armalytė, J., Skerniškytė, J., Bakienė, E., Krasauskas, R., Šiugždinienė, R., Kareivienė, V., Kerzienė, S., Klimienė, I., Sužiedėlienė, E., and Ružauskas, M. (2019). Microbial Diversity and Antimicrobial Resistance Profile in Microbiota From Soils of Conventional and Organic Farming Systems. Front Microbiol 10, 892. 10.3389/fmicb.2019.00892.
Krasauskas, R., Skerniškytė, J., Armalytė, J., and Sužiedėlienė, E. (2019). The role of Acinetobacter baumannii response regulator BfmR in pellicle formation and competitiveness via contact-dependent inhibition system. BMC Microbiol. 19, 241. 10.1186/s12866-019-1621-5.
Miškinytė, M., Juškaitė, R., Skerniškytė, J., Voldemarienė, V., Valuckas, K.P., Kučinskienė, Z.A., Sužiedėlis, K., and Sužiedėlienė, E. (2019). High incidence of plasmid-mediated quinolone resistance (PMQR) genes among antibiotic-resistant Gram-negative bacteria from patients of the Lithuanian National Cancer Center. Infect Dis (Lond) 51, 471–474. 10.1080/23744235.2019.1591637.
Skerniškytė, J., Krasauskas, R., Péchoux, C., Kulakauskas, S., Armalytė, J., and Sužiedėlienė, E. (2018). Surface-Related Features and Virulence Among Acinetobacter baumannii Clinical Isolates Belonging to International Clones I and II. Front Microbiol 9, 3116. 10.3389/fmicb.2018.03116.
Skerniškytė, J., Karazijaitė, E., Deschamps, J., Krasauskas, R., Armalytė, J., Briandet, R., and Sužiedėlienė, E. (2019). Blp1 protein shows virulence-associated features and elicits protective immunity to Acinetobacter baumannii infection. BMC Microbiol. 19, 259. 10.1186/s12866-019-1615-3.
Skerniškytė, J., Karazijaitė, E., Deschamps, J., Krasauskas, R., Briandet, R., and Sužiedėlienė, E. (2019). The Mutation of Conservative Asp268 Residue in the Peptidoglycan-Associated Domain of the OmpA Protein Affects Multiple Acinetobacter baumannii Virulence Characteristics. Molecules 24, 1972. 10.3390/molecules24101972.
Laboratory of Nucleic Acids Biochemistry (since 2020):
- Vepštaitė-Monstavičė I, Lukša-Žebelovič J, Apšegaitė V, Mozūraitis R, Lisicinas R, Stanevičienė R, Blažytė-Čereškienė L, Serva S, Servienė E. Profiles of Killer Systems and Volatile Organic Compounds of Rowanberry and Rosehip-Inhabiting Yeasts Substantiate Implications for Biocontrol. Foods. 2025 Jan 16;14(2):288. doi: 10.3390/foods14020288.
- Celitan E, Stanevičienė R, Servienė E, and Serva S (2024). Highly stable Saccharomyces cerevisiae L-BC capsids with versatile packing potential. Bioeng. Biotechnol. 12:1456453. doi: 10.3389/fbioe.2024.1456453.
- Vepštaitė-Monstavičė I, Lukša J, Strazdaitė-Žielienė Ž, Serva S, Servienė E. Distinct microbial communities associated with health-relevant wild berries. Environ Microbiol Rep. 2024 Dec;16(6):e70048. doi: 10.1111/1758-2229.70048.
- Konovalovas, A., Armalytė, J., Klimkaitė, L., Liveikis, T., Jonaitytė, B., Danila, E., Bironaitė, D., Mieliauskaitė, D., Bagdonas, E. & Aldonytė, R. (2024). Human nasal microbiota shifts in healthy and chronic respiratory disease conditions. BMC Microbiology, 24(1), 150. https://doi.org/10.1186/s12866-024-03294-5.
- Konovalovas, A., Armalytė, J., Klimkaitė, L., Liveikis, T., Jonaitytė, B., Danila, E., Bironaitė, D., Mieliauskaitė, D., Bagdonas, E. & Aldonytė, R. (2024). Insights into respiratory microbiome composition and systemic inflammatory biomarkers of bronchiectasis patients. Microbiology Spectrum, 12(12), e04144-23. https://doi.org/10.1128/spectrum.04144-23.
- Zebrauskiene, D., Sadauskiene, E., Dapkunas, J., Kairys, V., Balciunas, J., Konovalovas, A., Masiuliene, R., Petraityte, G., Valeviciene, N., Mataciunas, M., Barysiene, J., Mikstiene, V., Tomkuviene, M. & Preiksaitiene, E. (2024). Aortic disease and cardiomyopathy in patients with a novel DNMT3A gene variant causing Tatton-Brown–Rahman syndrome. Clinical Epigenetics, 16(1), 76. https://doi.org/10.1186/s13148-024-01686-y.
- Macedo DH, Grybchuk D, Režnarová J, Votýpka J, Klocek D, Yurchenko T, Ševčík J, Magri A, Dolinská MU, Záhonová K, Lukeš J, Servienė E, Jászayová A, Serva S, Malysheva MN, Frolov AO, Yurchenko V, Kostygov AY. Diversity of RNA viruses in the cosmopolitan monoxenous trypanosomatid Leptomonas pyrrhocoris. BMC Biol. 2023 Sep 12;21(1):191. doi: 10.1186/s12915-023-01687-y.
- Servienė E, Serva S. Recent Advances in the Yeast Killer Systems Research. Microorganisms. 2023 May 1;11(5):1191. https://doi.org/10.3390/microorganisms11051191.
- Merkys, A., Vaitkus, A., Grybauskas, A., Konovalovas, A., Quirós, M. & Gražulis, S. (2023). Graph isomorphism-based algorithm for cross-checking chemical and crystallographic descriptions. Journal of Cheminformatics, 15(1), 25. https://doi.org/10.1186/s13321-023-00692-1.
- Lingė, D., Gedgaudas, M., Merkys, A., Petrauskas, V., Vaitkus, A., Grybauskas, A., Paketurytė, V., Zubrienė, A., Zakšauskas, A., Mickevičiūtė, A., Smirnovienė, J., Baranauskienė, L., Čapkauskaitė, E., Dudutienė, V., Urniežius, E., Konovalovas, A., Kazlauskas, E., Shubin, K., Schiöth, H. B., Matulis, D. (2023). PLBD: protein–ligand binding database of thermodynamic and kinetic intrinsic parameters. Database, https://doi.org/10.1093/database/baad040.
- Lukša, J.; Celitan, E.; Servienė, E.; Serva, S. Association of ScV-LA Virus with Host Protein Metabolism Determined by Proteomics Analysis and Cognate RNA Sequencing. Viruses 2022, 14, 2345. https://doi.org/10.3390/v14112345.
- Grybchuk D, Procházková M, Füzik T, Konovalovas A, Serva S, Yurchenko V, Plevka P. Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly. Commun Biol. 2022 Aug 20;5(1):847. doi: 10.1038/s42003-022-03793-z.
- Saura A, Zakharova A, Klocek D, Gerasimov ES, Butenko A, Macedo DH, Servienė E, Zagirova D, Meshcheryakova A, Rogozin IB, Serva S, Kostygov AY, Yurchenko V. Elimination of LRVs Elicits Different Responses in Leishmania spp. mSphere. 2022 Aug 9:e0033522. doi: 10.1128/msphere.00335-22.
- Ravoityte, B.; Lukša, J.; Wellinger, R.E.; Serva, S.; Serviene, E. Adaptive Response of Saccharomyces Hosts to Totiviridae L-A dsRNA Viruses Is Achieved through Intrinsically Balanced Action of Targeted Transcription Factors. Fungi 2022, 8, 381. doi: 10.3390/jof8040381.
- Aitmanaitė L, Konovalovas A, Medvedevas P, Servienė E, Serva S. Specificity Determination in Saccharomyces cerevisiae Killer Virus Systems. Microorganisms. 2021 Jan 23;9(2):236. doi: 10.3390/microorganisms9020236.
- Ravoitytė B, Lukša J, Yurchenko V, Serva S, Servienė E. Saccharomyces paradoxus Transcriptional Alterations in Cells of Distinct Phenotype and Viral dsRNA Content. Microorganisms. 2020 Nov 30;8(12): E1902. doi: 10.3390/microorganisms8121902.
Molecular mechanisms of resistance to anticancer treatment:
Lysosome-targeted photodynamic treatment induces primary keratinocyte differentiation. Daugelaviciene N, Grigaitis P, Gasiule L, Dabkeviciene D, Neniskyte U, Sasnauskiene A. J Photochem Photobiol B. 2021 May;218:112183. doi: 10.1016/j.jphotobiol.2021.112183. Epub 2021 Mar 29. PMID: 33831753
Notch and Endometrial Cancer. Jonusiene V, Sasnauskiene A. Adv Exp Med Biol. 2021;1287:47-57. doi: 10.1007/978-3-030-55031-8_4. PMID: 33034025 Review.
Exogenous interleukin-1α signaling negatively impacts acquired chemoresistance and alters cell adhesion molecule expression pattern in colorectal carcinoma cells HCT116. Grigaitis P, Jonusiene V, Zitkute V, Dapkunas J, Dabkeviciene D, Sasnauskiene A. Cytokine. 2019 Feb; 114:38-46. doi: 10.1016/j.cyto.2018.11.031. Epub 2018 Dec 22. PMID: 30583087
Significance of Notch and Wnt signaling for chemoresistance of colorectal cancer cells HCT116. Kukcinaviciute E, Jonusiene V, Sasnauskiene A, Dabkeviciene D, Eidenaite E, Laurinavicius A. J Cell Biochem. 2018 Jul;119(7):5913-5920. doi: 10.1002/jcb.26783. Epub 2018 Apr 10. PMID: 29637602
Effect of mTHPC-mediated photodynamic therapy on 5-fluorouracil-resistant human colorectal cancer cells. Kukcinaviciute E, Sasnauskiene A, Dabkeviciene D, Kirveliene V, Jonusiene V. Photochem Photobiol Sci. 2017 Jul 1;16(7):1063-1070. doi: 10.1039/c7pp00014f. Epub 2017 May 16. PMID: 28509917
Molecular mechanisms of intracellular trafficking:
Kaipa JM, Starkuviene V, Erfle H, Eils R, Gladilin E. Transcriptome profiling reveals Silibinin dose-dependent response network in non-small lung cancer cells. PeerJ. 2020 Dec 16;8:e10373. doi: 10.7717/peerj.10373.
Liu, SJ; Majeed, W; Grigaitis, P; Betts, MJ; Climer, LK; Starkuviene, V; Storrie, B. Epistatic Analysis of the Contribution of Rabs and Kifs to CATCHR Family Dependent Golgi Organization. Front Cell Dev Bio, 2019, Aug 2;7:126. doi: 10.3389/fcell.2019.00126.
Starkuviene, V., Kallenberger, SM., Beil, N., Lisauskas, T., Schumacher, BS., Bulkescher, R., Wajda, P., Gunkel, M., Beneke, J., Erfle, H. (2019) High-Density Cell Arrays for Genome-Scale Phenotypic Screening. SLAS Discov. 2019 Mar;24(3):274-283. doi: 10.1177/2472555218818757.
Bulkescher, R., Starkuviene, V., & Erfle, H. (2017). Solid-phase reverse transfection for intracellular delivery of functionally active proteins. Genome Res. 2017 Oct;27(10):1752-1758. doi: 10.1101/gr.215103.
Gunkel, M., Erfle, H., & Starkuviene, V. (2016). High-Content Analysis of the Golgi Complex by Correlative Screening Microscopy. Methods Mol Biol, 111-121.
DEPARTMENT STAFF
|
Employee |
Position |
Contacts |
Courses taught |
List of Publications |
|
Head of Department, Associate Professor, Senior Researcher |
||||
| Dr. Lina Aitmanaitė | Assistant Professor, Researcher | |||
| Dr. Julija Armalytė | Professor, Senior Researcher | |||
| Jolanta Bagvilė | Junior Administrator |
|
||
| Dr. Enrika Celitan | Research Assistant, Lecturer | |||
| Gerda Jasinevičienė | Research Assistant | |||
| Andrius Jasinevičius | Research Assistant, Laboratory Technician, PhD Student | Saulėtekio al. 7, V237 kab. |
||
| Dr. Violeta Jonušienė | Associate Professor, Researcher | |||
| Laurita Klimkaitė | Research Assistant, Teaching Assistant | |||
| Dr. Aleksandras Konovalovas | Senior Researcher | |||
| Tomas Liveikis | Research Assistant, Laboratory Technician | |||
| Dr. Arvydas Markuckas | Associate Professor | |||
| Dr. Algirdas Mikalkėnas | Assistant Professor, Biologist | |||
| Ramutė Pagalytė | Senior Technician | |||
| Ignas Ragaišis | Teaching Assistant | |||
| Dr. Saulius Serva | Professor | |||
| Dr. Jūratė Skerniškytė | Senior Researcher | |||
| Dr. Vytautė Starkuvienė-Erfle | Professor | |||
| Dr. (HP) Edita Sužiedėlienė | Professor | |||
| Dr. (HP) Kęstutis Sužiedėlis | Professor | |||
| Dr. Aurelijus Zimkus | Biologists Researcher | |||
| Dr. Eglė Žalytė | Assistant Professor, Resarcher | (+370) 5 239 8229 Saulėtekio al. 7, V241 kab |
||
| Zigmantas Žitkus | Lecturer | |||
| Dr. Vilmantė Žitkutė | Resarcher, Assistant Professor | |||
| Visiting Staff | ||||
| Dr. Martina Rudgalvytė |
Assistant Professor (Brussels Free University, Belgium) |
|||
PARTNERS
Molecular Mechanisms of Bacterial Antibiotic Resistance and Pathogenesis:
- Dr. Irina Buchovec (Vilnius University, Faculty of Physics (The application of light technologies in biomedicine and food safety, inactivation of pathogens using antimicrobial photodynamic therapy)).
- Prof. Edvardas Danila (Vilnius University Hospital Santaros Klinikos, Centre of Pulmonology and Allergology (Human airway microbiome research)).
- Assoc. Prof. Silvija Kiverytė (Vilnius University Hospital Santaros Klinikos, Centre of Laboratory Medicine (Spread and evolution of antimicrobial resistance in the clinic)).
- Assoc. Prof. Agnė Kirkliauskienė (Vilnius University, Faculty of Medicine (Spread and evolution of antimicrobial resistance in the clinic)).
- Prof. Modestas Ružauskas (Lithuanian University of Health Sciences (The prevalence of antibiotic-resistant bacteria in the environment)).
Laboratory of Nucleic Acids Biochemistry:
- Dr. Elena Servienė (Nature Research Center, Vilnius)
- Dr. Vytautė Starkuvienė-Erfle (BioQuant, Heidelberg University, Heidelberg, Germany)
- Dr. Vyacheslav Yurchenko (Life Science Research Centre, University of Ostrava, Czech Republic)
Molecular mechanisms of resistance to anticancer treatment:
- Dr. Vida Časaitė (Institute of Biochemistry, Life Sciences Center, Vilnius University)
- Dr. Eglė Preikšaitienė (Institute of Biomedical Sciences, Faculty of Medicine, Vilnius University)
- Dr. Vytautė Starkuvienė (BioQuant, Heidelberg University, Heidelberg, Germany)
Molecular mechanisms of intracellular trafficking:
- Dr. R. Valiokas, FTMC
- Prof. Dr. Med. M. Keese, University Mannheim Clinics, Heidelberg University
- Prof. S. Serva, Vilnius University
- Dr. A. Sasnauskienė, Vilnius University
STUDY PROGRAMMES
Biochemistry
|
Biochemistry is the science that studies the chemical processes in living organisms and the fundamental principles of life based on these processes. We are proud that Vilnius University has been recognized as a leading institution in fostering the education of Lithuanian biochemists, who have contributed to the international recognition of our country's life sciences for over 60 years, since 1962. |
 |
|
If you want to study Biochemistry at the Master's level, apply to the Master's programme in Biochemistry at Vilnius University Life Sciences Center. Vilnius University Life Sciences Center also offers doctoral studies in Biochemistry. More information |
 |
Where and what do biochemists do?
- Scientists: Lithuanian and foreign research centers;
- Scientists and lecturers: Lithuanian and foreign universities;
- Project managers, specialists: international and Lithuanian life science, biotechnology, biopharmaceutical companies and start-ups;
- Specialists: medical, biological, and diagnostic laboratories;
- Project managers, specialists: high-tech equipment sales and service companies.
Molecular biology
|
Molecular Biology is a young field of science. Still, its history is full of amazing discoveries, and the impact of the knowledge and biotechnologies it is based on has a significant effect on our lives. The discovery of the structure of DNA, the fundamental molecule of life, in 1953, is considered the birth of molecular biology. One of the discoverers of the structure of DNA, Francis Crick, said, “Almost all aspects of life are engineered at the molecular level, and without understanding molecules, we can only have a very sketchy understanding of life itself. How does molecular biology differ from biochemistry? Biochemistry studies the chemical reactions that occur in living organisms. Biological molecules, such as enzymes, which serve as biological catalysts, participate in cellular chemical reactions. Molecular biology studies the structure of these molecules, how they interact with other molecules, and their location within the cell. Thus, biochemistry and molecular biology have much in common. How does molecular biology differ from biology? Biology studies biological species, their diversity, and the evolution of these species. However, knowledge and applications of molecular biology are also making their way into biology: today, using molecular biology methods, it is possible to precisely determine the identity of a species – something that was impossible a few decades ago. Why become a molecular biologist? The greatest challenge for molecular biologists in the 21st century is to unravel all the molecular processes occurring in the cells of various organisms. You can contribute to this challenge by choosing to study molecular biology at Vilnius University Life Sciences Center. |
 |
| "Molecules" – as molecular biology students are affectionately called – will be able to enjoy interesting lectures, internships, research in scientific laboratories, and a fulfilling student life. After completing their studies, molecular biology specialists will be welcome at life sciences research centers and academic institutions, as well as biotechnology, biopharmaceutical, and other high-tech companies, biomedical institutions, and science and technology parks.
If you want to study Molecular Biology at the Master's level, apply to the Molecular Biology Master's program at Vilnius University Life Sciences Center. If you want to pursue a doctoral degree, apply to Doctoral studies in Biochemistry or Biology at Vilnius University Life Sciences Center. |
 |
Where and what do molecular biologists do?
- Scientists: Lithuanian and foreign research centers;
- Scientists and lecturers: Lithuanian and foreign universities;
- Project managers, specialists: international and Lithuanian life science, biotechnology, biopharmaceutical companies and start-ups;
- Specialists: medical, biological, and diagnostic laboratories, food industry companies, environmental protection, public health, agriculture, and veterinary services;
- Project managers, specialists: high-tech equipment sales and service companies.
