|
MINDAUGAS ZAREMBA |
|
RESEARCH OVERVIEW
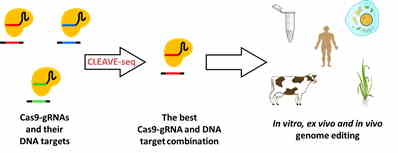
The recent development of CRISPR-Cas9 technology revolutionized genome editing field. Due to versatility to target almost any DNA sequence in the genome, CRISPR-Cas9 technology has a potential to be adopted for human genome editing therapy. Variety of studies demonstrated the possibility to modify human genome; however, one of the limits that the technology is facing to be adopted in clinic is its safety. It was demonstrated that various Cas9 nucleases are prone to cleave the DNA sites that are similar to the target sequences (off-target cleavage), resulting in the chromosome rearrangements or mutations causing cell death or even their transformation to cancer cells. Therefore, in order to make genome editing technology safer it is crucial to utilize a sensitive and reliable method for the detection of double-strand breaks (DSB) to evaluate the specificity of a selected DNA endonuclease in every particular case. The successful development of the technology is critical not only for fundamental research to detect and quantify DSBs induced using DNA endonucleases (CRISPR-Cas9 nucleases, zinc finger nucleases, TALEN nucleases, restriction endonucleases etc.), but also for institutions and companies like, to make the application of nuclease-based (including Cas9) technology safer for personalized genome editing and engineering in the clinic. Until now, there were various off-target detection methods published; however, they lack sensitivity, are experimentally complicated and require large coverage in sequencing what make them expensive. Furthermore, the lack of user-friendly protocols limits their applicability.
Our multidisciplinary team is working to develop a sensitive, cost-efficient, high-throughput and user-friendly DSB detection method that will allow determination of DNA endonuclease specificity for broad interest laboratories. The method will be adopted for Illumina and Oxford Nanopore Technologies sequencing platforms, making it available not only for the institutions containing high-throughput sequencing capabilities, but also for small laboratories reluctant to invest vast amount of finances for sequencing equipment and infrastructure. Developing the method we collaborated with Corteva Agricience, DuPont, and our method prototype CLEAVE-seq was successfully used in maize genome editing, resulting in a publication and a patent application (1). The DSB detection method is currently under further development (CPMA grant No. 01.2.2-CPVA-K-703-02-0010).
SELECTED PUBLICATIONS
- Young, J., Zastrow-Hayes, G., Deschamps, S., Svitashev, S., Zaremba, M., Acharya, A., Paulraj, S., Peterson-Burch, B., Schwartz, C., Djukanovic, V., Lenderts, B., Lanie Feigenbutz, L., Wang, L., Alarcon, C., Siksnys, V., May, G., Chilcoat, N. D., Kumar, S. CRISPR-Cas9 editing in maize: systematic evaluation of off-target activity and its relevance in crop improvement. Sci Rep. 2019 Apr 30, 9(1): 6729.
Methods for the Identification and Characterization of Double-Strand Break Sites and Compositions and Uses Thereof Patent application: WO/2019/217816, PCT/US2019/031719.