Project funder: Research Council of Lithuania
Grant no: S-MIP-23-127
Duration: 2023-2025
Amount awarded: €149899
Team (present/former): Skomantas Serapinas, Deimantė Stakelytė, Justina Gineitytė, dr. Eimanantas Ramonas, dr. Dalius Ratautas (PI)
Why it matters in one sentence
This project paves the way for handheld, low-cost sensors that spot tiny genetic variations in under an hour, so doctors and even in small clinics can customize treatment instantly.
Project overview
Future precision medicine centers on reading a patient's genetic code where care is delivered-quickly, inexpensively, and without bulky instruments. FindSNP is developing a plug and play electrochemical biosensor that detects single nucleotide polymorphisms (SNPs) in samples immediately after PCR, with no moving parts, temperature ramps or optics. By translating slight differences in DNA duplex stability into a real time electrical signal, the project aims to:
- Discriminate alleles in clinically actionable genes such as CYP2C19, which guides anti platelet and proton pump inhibitor therapy.
- Investigate operation on model samples and validate on saliva with minimal preparation.
- Detect unexpected mutations through thermodynamic analysis, laying groundwork for low cost electrochemical pseudo sequencing.
- Investigate electron transfer mechanisms at the DNA-electrode interface and possible strategies to amplify the typically weak DNA electrochemical signal.
Results – let’s get technical
- Signal‑amplified transducer. Designed a methylene‑blue / ascorbic‑acid redox cycle that tracks DNA hybridization on gold electrodes at one‑second resolution (target range 5–100 nM).
- Constant‑temperature SNP biosensor. Developed a cartridge‑ready assay in which a single PCR amplicon is interrogated by two allele‑specific reporters. As a proof of concept, the system discriminates CYP2C19 *1/*1, *1/*17 and *17/*17 diplotypes in untreated saliva within 80 minutes, showing concordance with sequencing.
- Thermodynamic mapping of unknown mutations. Calculated ΔΔG values distinguish single from double mismatches and highlight unknown variants, demonstrating the first step toward electrochemical pseudo‑sequencing of short loci.
- Outputs to date. One Q1 publication, one submitted manuscript, two conference presentations, one completed MSc thesis and an ongoing PhD project.
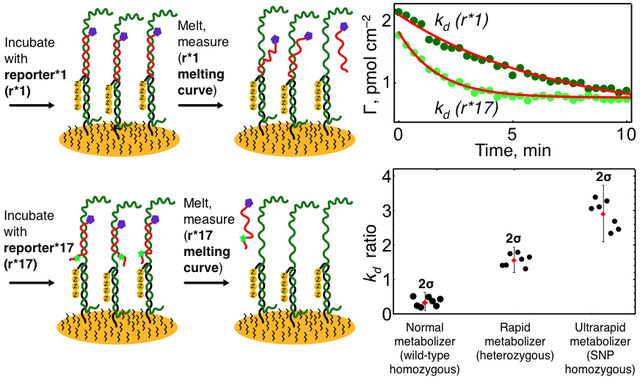
SNP sensor graphical abstract for CYP2C19 *1/*1, *1/*17 and *17/*17 genotyping. Sensor electrode is modified with ZNA-based probe, incubated with target (unknown) DNA and interrogated by two allele-specific reporters (r*1 and r*17). Melting kinetics are investigated resulting in different melting rates for reporters. Melting rate analyzes allow discrimination between diplotypes (*1/*1, *1/*17 and *17/*17) with high statistical significance.
Key publications
1. Justina Gineitytė, Skomantas Serapinas and Dalius Ratautas. Electrochemical DNA hybridization signal amplification system using methylene blue and ascorbic acid. Electrochimica Acta 2024, 507, 145146.
DOI: https://doi.org/10.1016/j.electacta.2024.145146
2. Skomantas Serapinas, Deimantė Stakelytė, Kornelija Tučinskytė, Miglė Tomkuvienė, Marius Dagys, and Dalius Ratautas. Constant temperature electrochemical biosensor for SNP detection in human genomic DNA based on DNA melting analysis. Submitted (2025).
Get in touch
For collaboration opportunities or further information, please contact PI dr. Dalius Ratautas ()
