Genome editing is one of the fastest-growing areas of modern biotechnology. Its capabilities are already transforming medicine and agriculture, and even enabling the revival of extinct animal species. However, effective and safe genome editing requires careful verification of the 'gene scissors' being used.
Researchers at Vilnius University’s Life Sciences Center (VU LSC) have developed an innovative method, CROFT-Seq, that allows scientists to precisely assess the efficiency and accuracy of CRISPR/Cas9 genome-editing tools directly in a test tube, significantly reducing research costs.
One of the method’s co-authors, Dr. Mindaugas Zaremba, Senior Researcher at the Protein–Nucleic Acid Interactions Laboratory of the VU Life Sciences Center’s Institute of Biotechnology, explains the patented method.
 |
| Dr. Mindaugas Zaremba. VU photo |
The Method Selects the Most Accurate 'Gene Scissors'
Dr. Zaremba explains that CROFT-Seq (CRISPR nuclease off-target detection by sequencing) is designed to determine the efficiency and accuracy of the most widely used genome-editing tools, such as CRISPR/Cas9 nucleases.
“This means that CRISPR/Cas9 nucleases – also known as ‘gene scissors’ – can be engineered in many different ways to cut a specific site in the genome. Conducting genome-editing experiments in living cells (in vivo) is very expensive, so it is beneficial to first test the selected ‘gene scissors’ in a test tube (in vitro) to find out how effectively they cut where they are supposed to, and whether they avoid cutting where they shouldn’t,” says the researcher.
The method allows scientists to select only the most efficient and accurate variants of 'gene scissors' for further experiments, thereby reducing the costs of subsequent research. Researchers first assess the cutting potential and precision of the 'gene scissors' in vitro before moving to other stages.
“Precision is crucial here, because when aiming to cure a specific genetic disease, one needs to cut exactly one or several specific genes at precise locations in the human genome to correct the mutations that cause the disease successfully. If these ‘gene scissors’ are used imprecisely, they can introduce double-strand DNA breaks in unintended locations, potentially leading to cell death or cancer. Our method helps prevent this by quickly and cost-effectively checking the efficiency and precision of the 'gene scissors' in vitro,” Dr. Zaremba explains.
A New Method Reduces Research Costs
According to Dr. Zaremba, although there are already many methods on the market with similar functions, thanks to an original strategy and unique solutions, the researchers were able to significantly speed up the process and reduce costs by up to tenfold.
“The CROFT-Seq method allows for very accurate studies while keeping costs low. That’s why it is attractive to both industry and research centers,” the scientist notes.
An equally important achievement is that simplifying the method has become suitable for automation using high-throughput robotic liquid-handling systems.
“We simplified the method to the point that practically all of its steps can be performed in a single test tube. Using robotic liquid-handling systems saves time and reduces the risk of errors,” Dr. Zaremba points out.
When choosing a method, analyzing the resulting data is also crucial. Many methods in this field use specific bioinformatic filters for sequencing data analysis, which can limit the interpretation of results.
“In our method, the use of filters is minimized. Therefore, if the ‘gene scissors’ cut DNA not only at their target site (on-target) but also elsewhere (off-target), our method detects such inaccurate cuts. In contrast, the filters used in other methods may inadvertently exclude a genuine off-target site, distorting the final results,” says Dr. Zaremba.
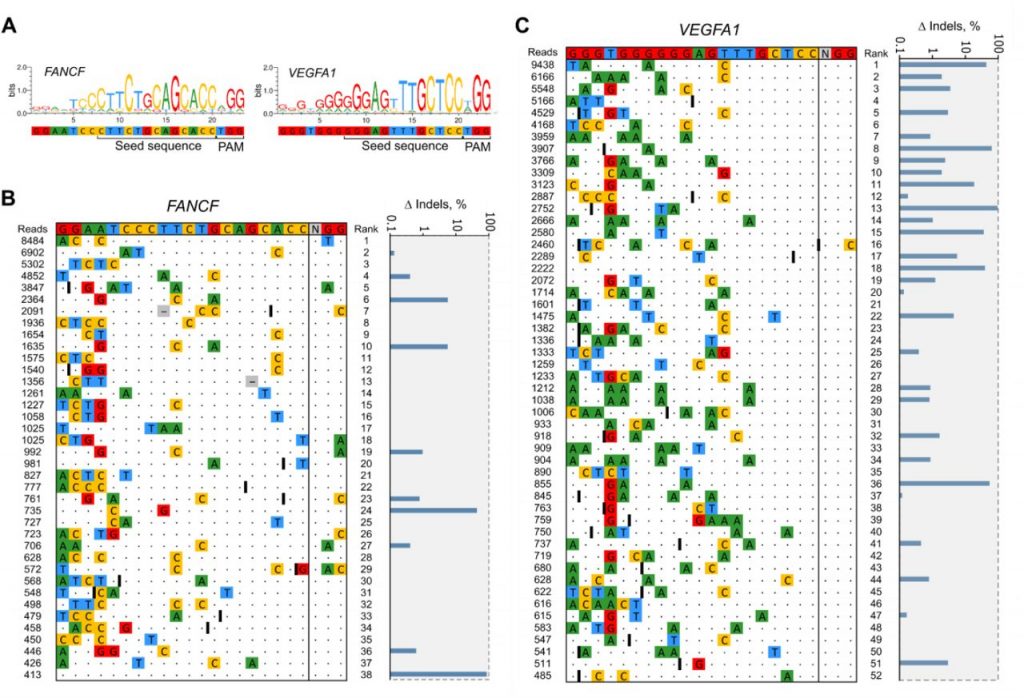 |
| Detection of off-target sites using the CROFT-Seq method. |
We Must Make an Effort to Catch up with the U.S.
Dr. Zaremba and his colleagues are currently carrying out a project funded by the VU Science Promotion Fund titled Platform for Assessing the Precision of Various Genome-Editing Tools.
The team aims to expand CROFT-Seq to other increasingly popular genome-editing tools, such as CRISPR/Cas12 or TnpB enzymes, as well as base editors.
“We want to increase the flexibility and appeal of our method by enabling the study of other genome-editing tools. We developed the method while implementing a project funded by the Central Project Management Agency – Double-Strand DNA Break Sequencing Center,” says the researcher.
The team decided to patent the method and has filed applications to protect it in the European, U.S., and Japanese markets.
“These markets are the most advanced in genome-editing research, and the U.S. is also a leader in applying research results. The U.S. has simpler regulatory mechanisms than the European Union (EU), so the EU is lagging. In this race, we are losing to the U.S. and missing significant opportunities for applying CRISPR/Cas technologies in agriculture, animal husbandry, and human healthcare,” the scientist observes.
In 2023, the first CRISPR/Cas9-based medicine was approved for treating sickle cell anemia. This allowed for the successful treatment of this monogenic disease and marked a true breakthrough. Numerous preclinical studies are underway to apply this technology to treat other diseases.
“Many genetic diseases are not monogenic. This means that their occurrence is caused by mutations in several genes, creating much greater challenges in curing them. I believe that CRISPR/Cas technology will make significant progress in treating such diseases over time,” Dr. Zaremba says.
Another example of genome editing applications is the revival of extinct animal species – already being undertaken by private companies in the U.S. “This only further proves the potential of the technology. Of course, there may be considerable safety and ethical challenges, but I hope they can be resolved,” the researcher adds.
More information about the patented invention can be found here. If you are interested in licensing this invention, please contact:
