|
VYTAUTAS SMIRNOVAS
|
|
RESEARCH OVERVIEW
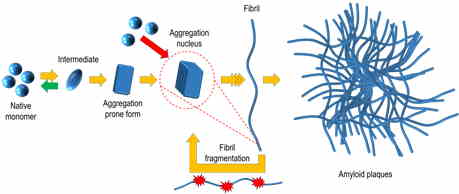
Protein misfolding and aggregation into amyloid structures is involved in many diseases, including such neurodegenerative disorders as Alzheimer’s and Parkinson’s, systemic amyloidoses and even some localized diseases such as type II diabetes or cataract. There is increasing evidence of the amyloid nature of proteinaceous infectious particles – prions. One of the possible ways of prion spreading is a self-replication of amyloid-like fibrils; thus, there is a chance of all amyloid-associated diseases to be potentially infective.
The ability of the same protein to adopt distinct pathogenic conformations was first reported in studies of infectious prions, and such conformations were referred to as strains. Strain-like polymorphism was reported for several other amyloid proteins. It highly increases the complexity of disease mechanisms and may be one of the reasons for the slow progress in drug research.
Our team studies the effects of environmental factors such as temperature, pressure, intensity and type of agitation, pH, ions, macromolecular crowding and the presence of different organic solvents, ligands and biomolecules on aggregation kinetics, thermodynamic stability and the structural properties of amyloid-like fibrils. We believe that only comprehensive knowledge of all factors may provide a genuine understanding of the mechanisms of amyloid self-replication, complexity of fibril polymorphism and lead towards curing amyloid-related diseases.
We are interested in comparing the aggregation profiles of different proteins and testing possibilities of their co-aggregation. The group has experience in the expression and purification of recombinant amyloid beta, alpha-synuclein, different isoforms of full-length Tau proteins, a variety of mammalian prion proteins (derived from different species and with different mutations), S100A9 protein, superoxide dismutase, sup35NM domain and beta-microglobulin. The main methods used to follow amyloid formation include UV, visible and fluorescence spectrometry (a Thioflavin T fluorescence assay as the main method to follow kinetics), Fourier transform infrared spectrometry and atomic force microscopy.
SELECTED PUBLICATIONS
- Ziaunys, M., Sakalauskas, A., Smirnovas, V. Identifying insulin fibril conformational differences by thioflavin-T binding characteristics. Biomacromolecules. 2020, 21(12): 4989–4997.
- Pansieri, J. et. al. Templating S100A9 amyloids on Aβ fibrillar surfaces revealed by charge detection mass spectrometry, microscopy, kinetic and microfluidic analyses. Chemical Science. 2020, 11(27): 7031–7039.
- Ziaunys, M., Sneideris, T., Smirnovas, V. Self-inhibition of insulin amyloid-like aggregation. Physical Chemistry Chemical Physics. 2018, 20(43): 27638–27645.
- Kim, C. et. al. Artificial strain of human prions created in vitro. Nature Communications. 2018, 9(1): 2166.
- Sneideris, T., Darguzis, D., Botyriute, A., Grigaliunas, M., Winter, R., Smirnovas, V. PH-driven polymorphism of insulin amyloid-like fibrils. PloS one. 2015, 10(8).
RESEARCH ACTIVITIES in 2020
Polymorphism of Amyloid Fibrils
Thioflavin T (ThT) fluorescence assay is one of the main methods used to detect amyloid fibrils and follow aggregation kinetics. We have demonstrated that deeper studies of ThT binding and fluorescence characteristics makes it possible to differentiate between structurally distinct amyloid aggregates (Ziaunys et. al. Biomacromolecules. 2020) without employing more complex techniques. Using this knowledge, we were able to perform analysis of many samples in a short time and found out that distinct conformations of prion protein amyloid fibrils can form even in the identical conditions (Ziaunys et. al. Scientific Reports. 2020).