|
VIRGINIJUS ŠIKŠNYS |
|
RESEARCH OVERVIEW
In recent years, Cas9 has revolutionized the genome-editing field and enabled a broad range of applications from basic biology to biotechnology and medicine. Cas9 can be guided to specific locations within complex genomes by a short RNA search string. Using this system, DNA sequences within the endogenous genome and their functional outputs are now easily edited or modulated in virtually any organism of choice.
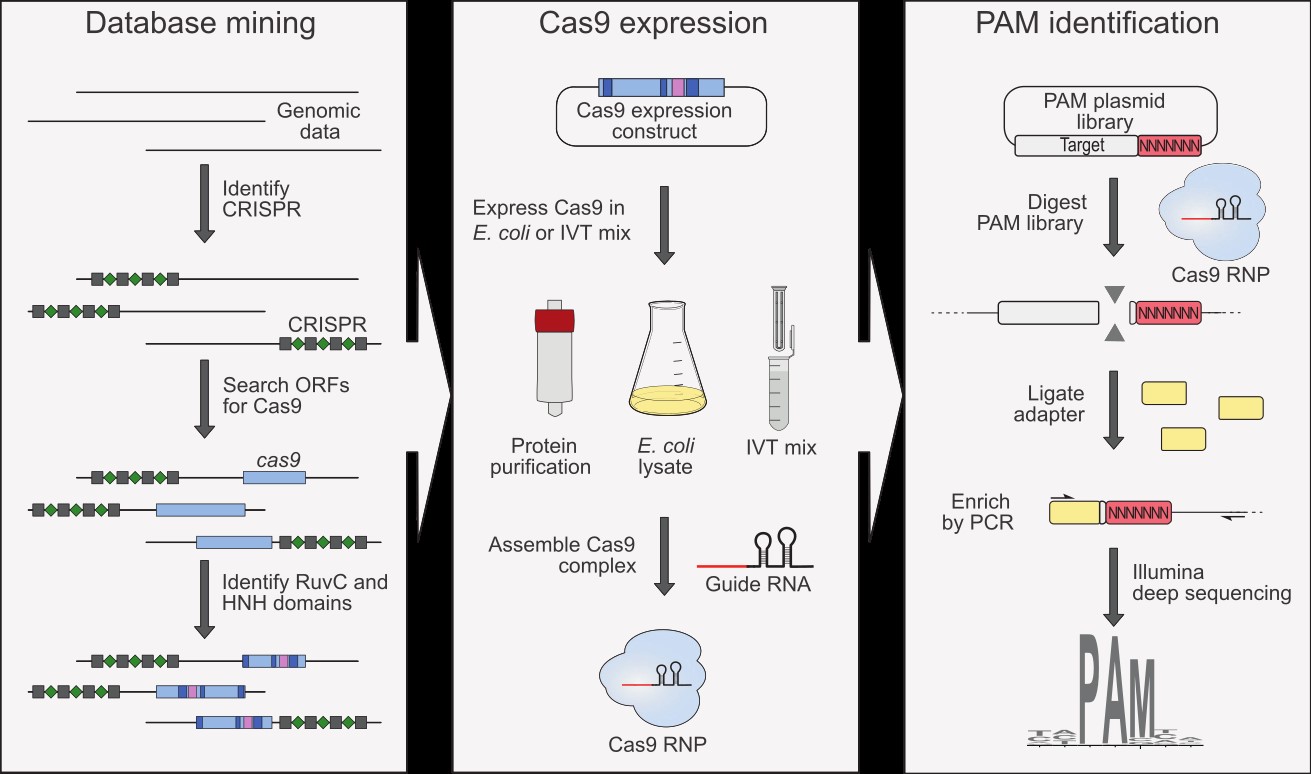
Cas9 specificity is dictated by base pairing of the guide RNA to the complementary DNA strand, however to initiate hybridization, a short protospacer adjacent motif (PAM) sequence is required in the vicinity of the target sequence. The PAM is recognized by the Cas9 protein and varies among Cas9s. To characterize the PAM recognition diversity provided by Cas9 orthologs, we developed a phylogeny-guided bioinformatics approach and streamlined our experimental procedures for Cas9 expression and RNP complex assembly using cell lysates and in vitro translation mixtures. This approach could be easily adapted for the characterization of other CRISPR-Cas nucleases that require PAM sequences and generate double-strand breaks following target recognition (Karvelis et al. Methods Enzymol. 2019, 616: 219–240).
SELECTED PUBLICATIONS
- Gasiunas, G., Barrangou, R., Horvath. P., Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012, 109: E2579–86.
- Šikšnys, V., Gasiūnas, G., Karvelis, T. RNA-directed DNA cleavage by the Cas9-crRNA complex. US patent. 9 637 739 2017.05.02
- Muller, M., Lee, C. M., Gasiunas, G., Davis, T. H., Cradick, T. J., Siksnys, V., Bao, G., Cathomen, T., Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Therapy. 2016, 24: 636–644.
- Karvelis, T., Gasiunas, G., Young, J., Bigelyte, G., Silanskas, A., Cigan, M. and Siksnys, V. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biology. 2015, 16: 253.
- Karvelis, T., Young, J. K., Siksnys, V. A pipeline for characterization of novel Cas9 orthologs. Methods Enzymol. 2019, 616: 219–240.
RESEARCH ACTIVITIES in 2020
Catalogue of Biochemically Diverse CRISPR-Cas9 Orthologues
Bacterial Cas9 nucleases from type II CRISPR-Cas antiviral defence systems are very diverse. We combined a bioinformatic analysis and biochemical screen to explore this largely uncharacterized diversity. We applied cell-free biochemical screens to assess the PAM and guide RNA requirements of 79 Cas9 proteins, thus identifying at least 7 distinct gRNA classes and 50 different PAM sequence requirements. PAM recognition spans the entire spectrum of T-, A-, C-, and G-rich nucleotides, from single nucleotide recognition to sequence strings longer than 4 nucleotides. Characterization of a subset of Cas9 orthologues using purified components reveals additional biochemical diversity, including both narrow and broad ranges of temperature dependence, staggered-end DNA target cleavage, and a requirement for long stretches of homology between gRNA and DNA target. Our results expand the available toolset of RNA-programmable CRISPR-associated nucleases (Gasiunas et al., Nat Commun. 2020, 11: 5512; Wang et al. CRISPR J. 2020, 3(6): 427–430).
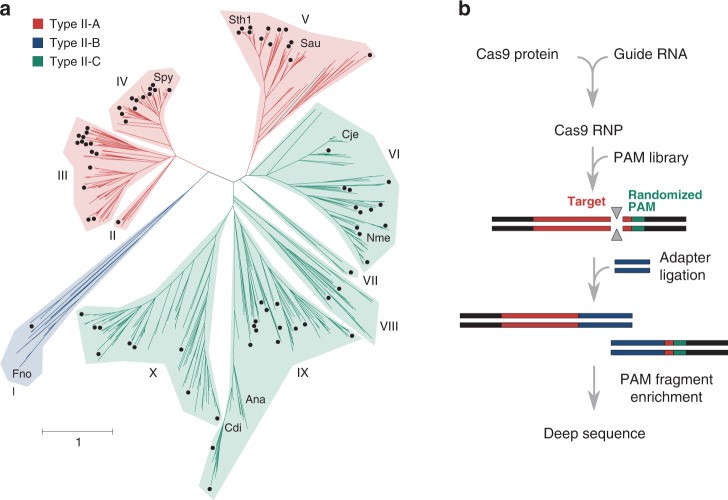
Characterization of Cas9 diversity
Characterisation of Compact CRISPR-Cas12f Nuclease
Over the past several years, Cas9 and Cas12 endonucleases have been adopted as robust genome editing and transcriptome manipulation tools. Although both nucleases have been widely used, the size of Cas9 and Cas12 provides constraints on cellular delivery that may limit certain applications, including therapeutics. Recently, small CRISPR-associated effector proteins (Cas12f) belonging to the type V-F subtype have been identified through the mining of sequence databases. The majority of Cas12f proteins are nearly half the size of the smallest Cas9 or Cas12 nucleases.
We selected a collection of 10 exceptionally compact (422-603 amino acids) CRISPR-Cas12f nucleases that recognize and cleave dsDNA in a PAM dependent manner. Categorized as class 2 type V-F, they originate from the previously identified Cas14 family and distantly related type V-U3 Cas proteins found in bacteria. Using biochemical methods, we demonstrate that a 5' T- or C-rich PAM sequence triggers dsDNA target cleavage. Based on this discovery, we evaluated whether they can protect against invading dsDNA in Escherichia coli and find that some but not all can. Altogether, our findings show that miniature Cas12f nucleases can protect against invading dsDNA like much larger class 2 CRISPR effectors and have the potential to be harnessed as programmable nucleases for genome editing (Karvelis et al., Nucleic Acids Res. 2020, 48(9): 5016–5023).
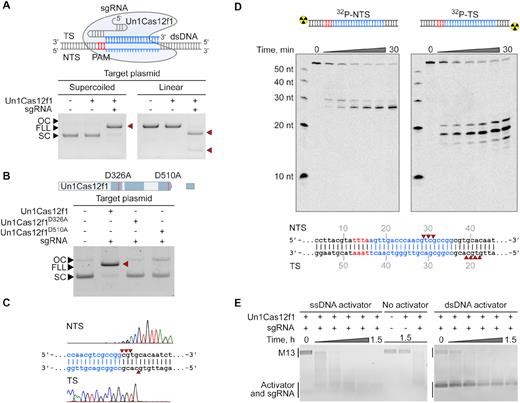
Un1Cas12f1 (Cas14a1) RNP complex is a PAM-dependent dsDNA endonuclease
Workpackage 4: Alternative Genome Editing Tools for the Retina
Over the past years, Cas9 and Cas12a nucleases have been adopted as robust genome editing and transcriptome manipulation tools. To recognize and cleave a dsDNA target, both Cas9 and Cas12 require a short sequence, termed the protospacer adjacent motif (PAM), in the vicinity of a DNA sequence targeted by the gRNA. The PAM requirement imposes a serious bottleneck in therapeutic editing where often a precise targeting close to the mutation site is required. If there is no PAM near the target site, Cas9 and Cas12a proteins cannot access it. Next, there are difficulties with the delivery of Cas9 and Cas12a into tissues or cells to achieve therapeutic effects. AAVs often would be a preferable choice, however, these vectors have their own cargo packaging limit, thus small size Cas nuclease variants are highly desirable. Here we aim to assess the therapeutic potential of novel Cas9 orthologues and variants that cut close to the desired mutation sites in EYS and USH2A, and explore the gene editing potential of the miniature Cas12f nucleases in HEK293 cells. The most promising Cas nucleases will be selected and further optimized in either wild type, or patient- derived retinal organoids, in collaboration with partners within the consortium.

Project partners: NL, IT, SW, BE, DE, FR



