Genetic and Structural Diversity of Bacteriophages
Bacteriophages (phages), the viruses that infect bacteria, are probably the most numerous biological entities on the planet, and they are also exceptionally diverse. Despite the fact that phages as model organisms have featured in many of the key studies of the last century and basically have helped transform biology into a modern science, they remain to be of great significance both in fundamental and applied research. For example, to combat the ever-growing antibiotic resistance in bacteria, a variety of promising phage-inspired antibacterial approaches as well as innovative techniques based on phage-borne enzymes (e.g., lysins) or structural proteins (e.g., tail spike/fibre) are being developed. The results obtained while studying unique phages isolated from different ecosystems by the scientists of the Department of Molecular Microbiology and Biotechnology show that the diversity of phages, in terms of virion structure, physiology and genetics, is enormous, and that we have not even begun to properly harvest it. In fact, every single phage studied not only provides novel insights into the nature of bacterial viruses, but can also be used as a source of novel building blocks for the construction of multifunctional nanomaterials or can be exploited in the detection/biocontrol of pathogenic bacteria
The phage group of the Department of Molecular Microbiology and Biotechnology has long focused on the isolation and molecular analysis of novel phages with unique structure, host range or physiology. Over the last five years, researchers of the Department carried out ten different projects funded by Vilnius University (MSF-LMT-2) and the Research Council of Lithuania (MIP-002/2014, P-MIP-19-259, 01.2.2-LMT-K-718-03-0099, 01.2.2-LMT-K-718-01-0019). Several projects were conducted in collaboration with the research groups of the Institute of Biotechnology (S-MIP-17-47), the Nature Research Centre (P-MIP-17-6, P-MIP-20-256) and the Institute of Biosciences (SIT-7/2015), as well as Kyoto University (S-LJB-17-1). The aims of the projects ranged from the investigation of gene expression profiles of novel bacteriophages, and elaboration of new systems for genome engineering of lytic bacteriophages to the investigation of molecular mechanisms of adaptation of low-temperature viruses to the mesophilic host. A number of unique Pantoea, Arthrobacter, and Buttiauxella phages have been isolated, characterised and published in the process [1–5]. In total, seventeen peer-reviewed scientific publications have been published by the research group during the last five-years.
SELECTED PUBLICATIONS:
- Noreika, A., Meškys, R., Lazutka, J., Kaliniene, L. Complete genome sequence of Buttiauxella phage vBButMGuL6. Arch Virol. 2020, 165: 2685–2687.
- Truncaitė, L., Šimoliūnienė, M., Alijošius, L., Petrauskaitė E., Kiaušaitė L., Meškys R., Skapas M., Šimoliūnas, E. Complete genome analysis of Pantoea agglomerans-infecting bacteriophage vBPagMAAM22. Arch Virol. 2020, doi: 10.1007/s00705-020-04705-4.
- Šimoliūnienė, M., Truncaitė, L., Petrauskaitė, E., Zajančkauskaitė, A., Meškys, R., Skapas, M., Kaupinis, A., Valius, M., Šimoliūnas, E. Pantoea agglomerans-infecting bacteriophage vBPagSAAS21: a cold-adapted virus representing a novel genus within the family Siphoviridae. Viruses. 2020, 12(4): 479. doi: 10.3390/v12040479.
- Šimoliūnas, E., Truncaitė, L., Rutkienė, R., Povilonienė, S., Goda, K., Kaupinis, A., Valius, M., Meškys, R. The robust self-assembling tubular nanostructures formed by gp053 from phage vBEcoMFV3. Viruses. 2019, 11(50). doi.org/10.3390/v11010050.
- Kaliniene, L., Šimoliūnas, E., Truncaitė, L., Zajančkauskaitė, A., Nainys, J., Kaupinis, A., Valius, M., Meškys, R. Molecular analysis of Arthrobacter myovirus vB_ArtM-ArV1: we blame it on the tail. Journal of Virology. 2017, 91(8): e00023.
Ecogenomics and Potential Biogeochemical Impacts of Viruses in Gypsum Karst Lake Ecosystems
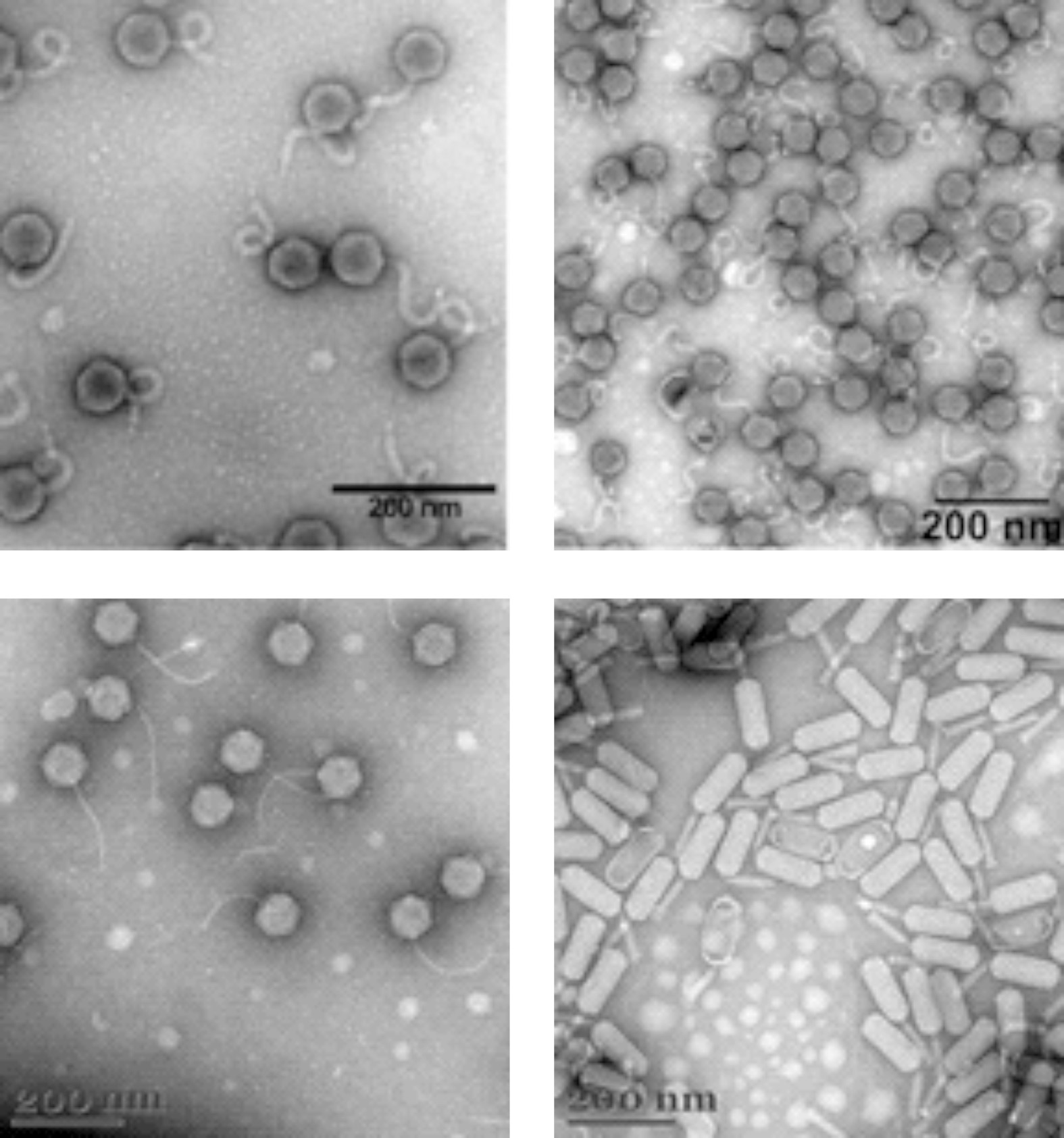
In this study, we present a number of bacteria and bacteriophages isolated from water samples of the unique sulfate-type gypsum karst lakes Kirkilai and Ramunėlis located near Biržai, Lithuania.
Nine bacteriophages were isolated using Aeromonas, Bacillus, Paracoccus, Pseudomonas, Pararheinheimera and Pseudaeromonas as the host for phage propagation and phage growth experiments. TEM analysis revealed that most of the phages are myoviruses, Aeromonas phage KLEA5, Bacillus phages KLEB27-1 and KLEB30-3S are siphoviruses, while Paracoccus phage KLEP18-1 is a podovirus. Most of aforementioned phages have isometric heads of about 60–65 nm in diameter with an exception of Bacillus phage KLEB27-3, which has a head about 108 nm in diameter and a contractile tail about 285 nm long. Phylogenetic analysis revealed that the vast majority of the phages have no close phylogenetic relatedness to other viruses published to date and potentially represent new genera within the order Caudovirales.
The data presented in this study will not only expand our knowledge of the diversity of bacteriophages but will also lead to a better understanding of almost unexplored communities of bacteria and viruses in the unique sulfate-type gypsum karst lakes (Grant Nr. MSF-LMT-2).
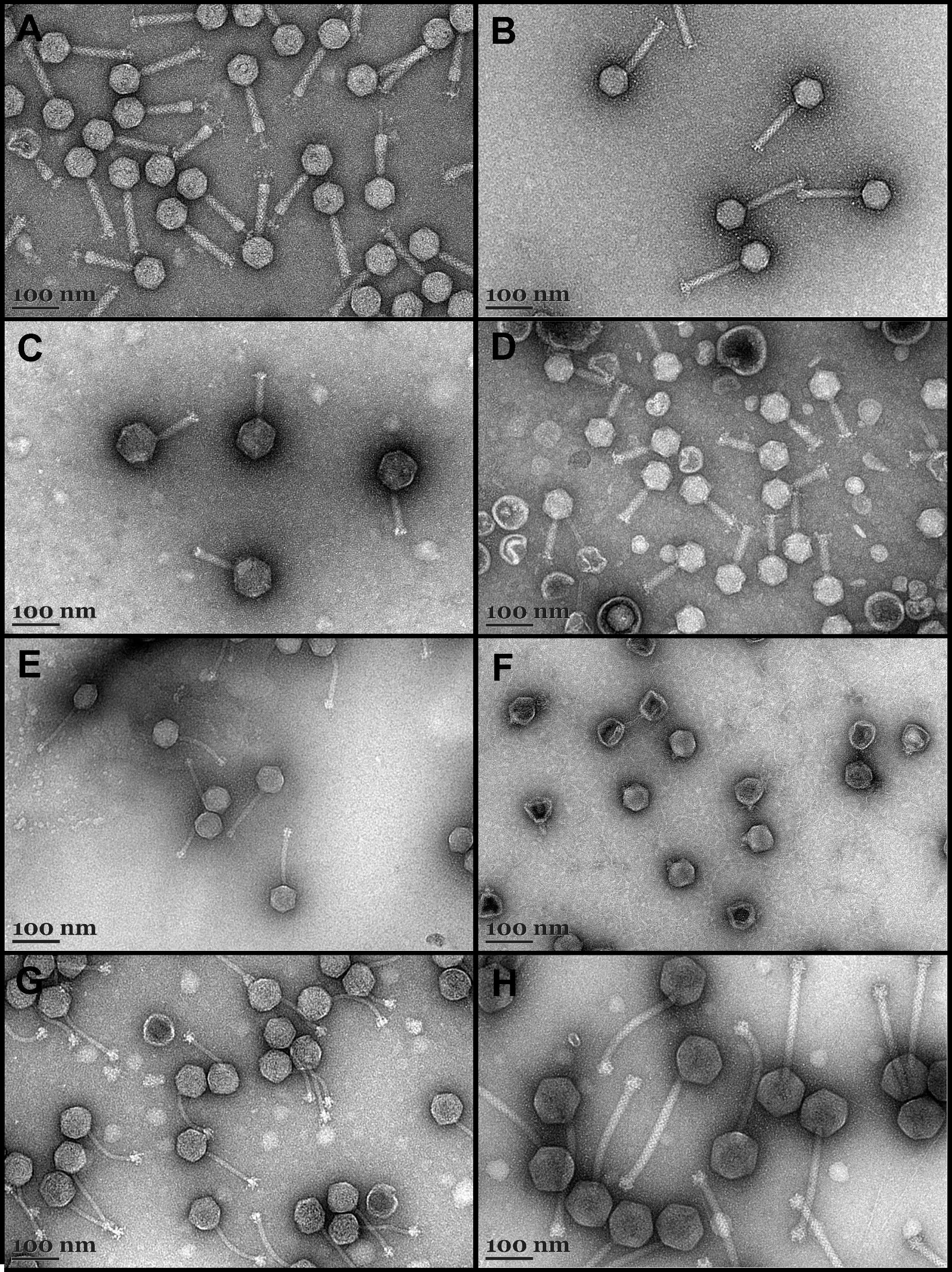
Transmission electron micrographs of phage particles isolated from the gypsum karst lakes Kirkilai and Ramunėlis: KLER1-1 (A), KLER1-2 (B), KLEA5 (C), KLEP7 (D), KLEP17-4 (E), KLEP18-1 (F), KLEB27-1 (G) and KLEB27-3 (H).
Complete Genome Sequence of Buttiauxella Phage vB_ButM_GuL6
Bacteriophage vB_ButM_GuL6 is the first virus isolated from Buttiauxella. Electron microscopy revealed that vB_ButM_GuL6 belongs to the family
Myoviridae, order Caudovirales. The genome of vB_ButM_GuL6 is a linear, circularly permuted 178,039-bp dsDNA molecule with a GC content of 43.4%. It has been predicted to contain 282 protein-coding genes and two tRNA genes, tRNA-Met and tRNA-Gly. Using bioinformatics approaches, 99 (36%) of the vB_ButM_GuL6 genes were assigned a putative function. Genome-wide comparisons and phylogenetic analysis indicated that vB_ButM_GuL6 represents a new species of the subfamily Tevenvirinae and is most closely related to Escherichia coli virus RB43. These phages, together with Cronobacter phages Miller, CfP1, and IME-CF2, likely form a new genus within the subfamily Tevenvirinae.
(Noreika et al. Arch Virol. 2020, 165: 2685–2687).

Functional genome map of bacteriophage GuL6
The colour code is as follows: blue, structural proteins and those involved in virion morphogenesis; yellow, DNA replication, recombination, and repair; green, transcription, translation, and nucleotide metabolism; red, lysis/phage-host interactions; black, genes of unknown function; white, genes that encode unique proteins with no reliable homology to database entries; pink, tRNAs