|
|
NOMEDA KUISIENĖ
|
Prokaryotes represent the largest source of biotechnologically relevant products in nature. New species of prokaryotes are continuously described, and new strains of the “old” species are also continuously isolated. It is known that every new bacterial strain adds dozens of new genes to the genome of its own species, and at least some of these new genes can be exploited for the development of novel, biotechnologically relevant products.
Prokaryotes developed a range of enzymes that degrade polysaccharides, producing oligosaccharides. Different bioactivities useful for human health were reported for oligosaccharides; they are also used as prebiotics in functional food. The enzymatic production of these compounds is the most promising.
Prokaryotes also developed a whole range of structural proteins, and some of them (collagen-like proteins, for example) can be used for the construction of biomaterials with the desirable properties for regenerative medicine.
Most bacteria produce antimicrobial compounds of different nature: volatile compounds, bacteriocins, antibiotics. In practice, they can be used for both the prevention and treatment of infections. Screening for novel antimicrobial compounds is regarded to be the most promising strategy for overcoming the problem of antimicrobial resistance.
SELECTED PUBLICATIONS
- Lukoseviciute, L., Lebedeva, J., Kuisiene, N. Diversity of polyketide synthases and nonribosomal peptide synthetases revealed through metagenomic analysis of a deep oligotrophic cave. Microbial Ecology. 2020, in press.
- Kananavičiūtė, R., Kvederavičiūtė, K., Dabkevičienė, D., Mackevičius, G., Kuisienė, N. Collagen-like sequences encoded by extremophilic and extremotolerant bacteria. Genomics. 2020, 112: 2271–2281.
- Bukelskis, D., Dabkeviciene, D., Lukoseviciute, L., Bucelis, A., Kriaučiūnas, I., Lebedeva, J., Kuisiene, N. Screening and transcriptional analysis of polyketide synthases and non-ribosomal peptide synthetases in bacterial strains from Krubera–Voronja Cave. Frontiers in Microbiology. 2019, 10: 2149.
- Kirtikliene, T., Naugzemys, D., Steponkiene, A., Bogdevic, R., Vizuje, G., Zvingila, D., Kusiene, N. Evaluation of the inter- and intrahospital spread of multidrug resistant Gram-negative bacteria in Lithuanian hospitals. Microbial Drug Resistance. 2019, 25: 326–335.
- Tratulyte, S., Miciuleviciene, J., Kuisiene, N. First genotypic characterization of toxigenic Clostridioides difficile in Lithuanian hospitals reveals the prevalence of the hypervirulent ribotype 027/ST1. European Journal of Clinical Microbiology & Infectious Diseases. 2019, 38: 1953–1959.
Biosynthesis Genes of Bioactive Compounds: Evaluation of Diversity and Expression Analysis in the Unique Environment
The identification of novel compounds with antibacterial, antifungal, anticancer, antiviral, antidiabetic, antiprotozoal and other bioactivities represents an important field of modern biomedical research. Microorganisms are the main targets in this research because of their high potential to produce these bioactive compounds. Bioactive compounds can be difficult to identify phenotypically because of a few reasons: the amount of these compounds can be beyond the detection limits; certain experimental conditions can be inappropriate for the induction of the biosynthesis of these compounds; the coding genes of bioactive compounds can be silent etc. The problem can be solved, and the real potential of bioactivity can be determined through the analysis of biosynthesis genes and not via that of the bioactive compounds themselves. The aim of the current project is to reveal the diversity and prevalence of bioactive compound biosynthesis genes in the bacteria of the deepest cave of the Earth, the Krubera-Voronja Cave. Polyketide synthase, nonribosomal peptide synthetase and bacteriocin biosynthesis genes were under investigation in this project (Lukoseviciute et al. Microbial Ecology. 2020;
Bukelskis et al. Frontiers in Microbiology. 2019).
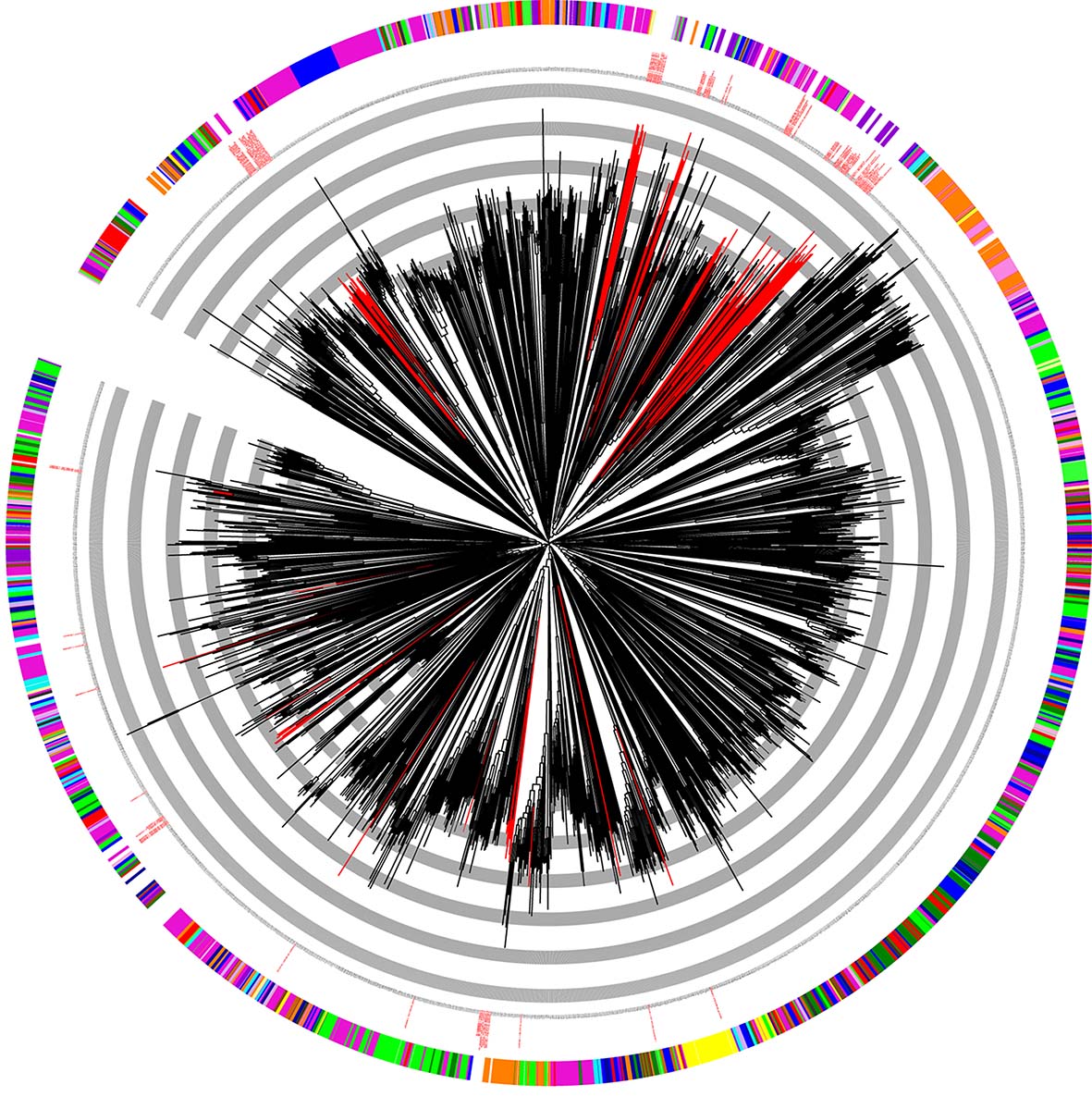
Fig. 1. Phylogenetic diversity of the adenylation domains of the nonribosomal peptide synthetases in the metagenome of Krubera-Voronja Cave.
Identification, Expression and Characterization of Bacterial Collagen-Like Proteins
During the last decade, a large number of collagen-like proteins have been identified in bacteria mainly through an in silico analysis. Only a few bacterial collagen-like proteins have been expressed in Escherichia coli. It was shown that these recombinant bacterial proteins adopt a classical triple-helix conformation and exhibit high thermal stability. The amino acid composition of bacterial collagen-like proteins varies from species to species, and from protein to protein, conferring the different characteristics to these proteins. Collagen-like proteins can be produced in large quantities by recombinant methods, and the construction of proteins with the desirable characteristics can also be carried out. Therefore, bacterial collagen-like proteins represent an excellent source for the design of new biomaterials with the desirable structural properties and functions. The identification, expression and characterization of bacterial collagen-like proteins represent a highly attractive and important area of research work in the fields of regenerative medicine and biotechnology (Kananavičiūtė et. al. Genomics. 2020).
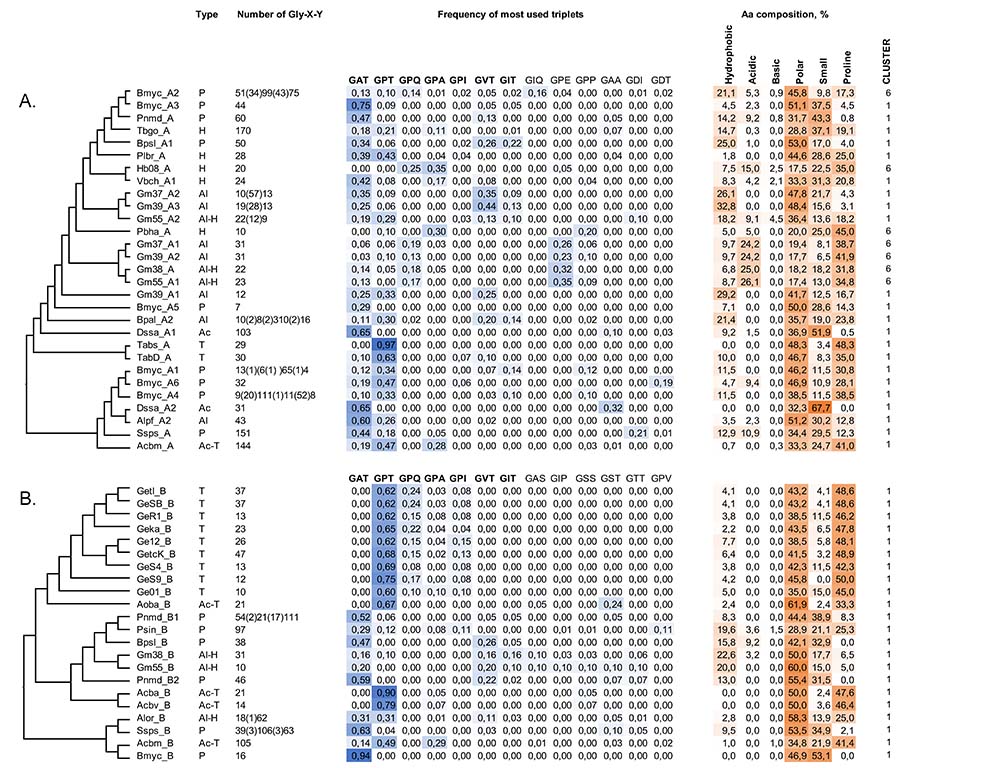
Fig. 2. Collagen-like sequence proteins having BclA_C (A) and Exospore_TM domains (B).
Molecular Epidemiology of Pathogenic Bacteria in Lithuanian Healthcare Institutions
Bacterial resistance to antimicrobial agents plays an important role in healthcare institutions nowadays. Spread of multidrug resistant bacteria can occur during inter- and intra-hospital transmissions among patients and hospital personnel. One of the highest rates of resistance in healthcare institutions is observed in Acinetobacter spp. isolates, which causes outbreaks around the world and is highly adaptable to changes both in the environment and in the use of antibiotics. These characteristics lead to a high rate of occurrence of multidrug-resistant Acinetobacter spp. cases in the environment.
Methods of molecular epidemiology, such as virulence factors determination, resistance genes distribution and genotyping, are used to better understand the antimicrobial resistance patterns of Acinetobacter spp. and can be used to strengthen the control of multidrug resistant infections in healthcare institutions and to prevent potential outbreaks of this pathogen in the future
(Kirtikliene et al. Microbial Drug Resistance. 2019;
Tratulyte et al. European Journal of Clinical Microbiology & Infectious Diseases. 2019).
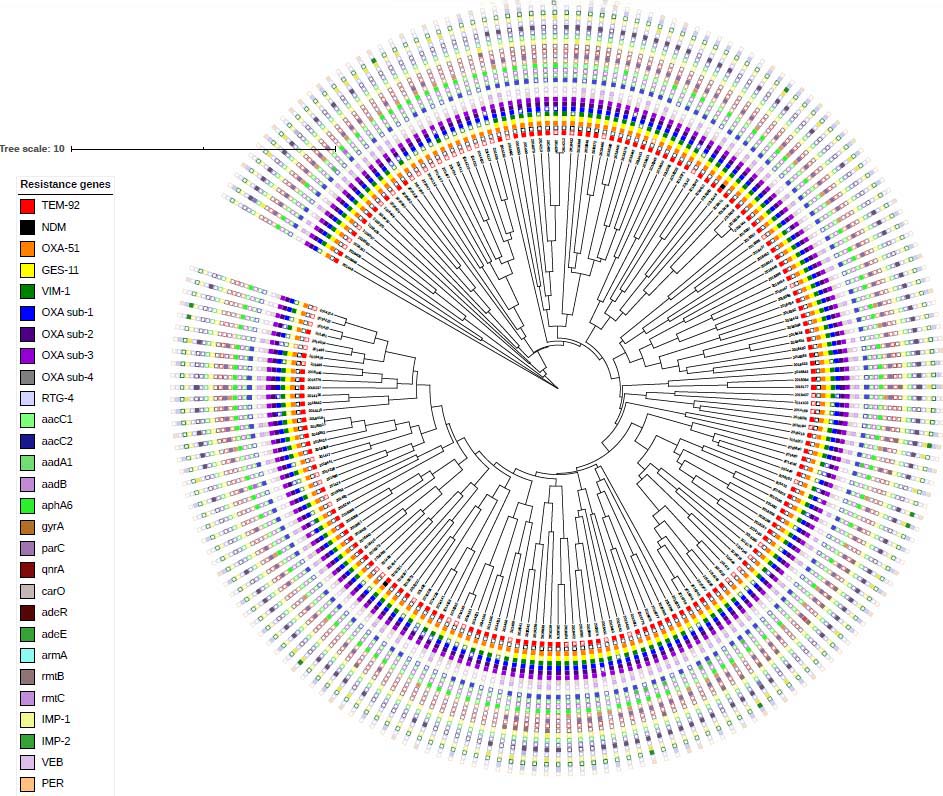
Fig. 3. Dendrogram for the BOX-PCR pattern of Acinetobacter spp. isolates resistant to antibiotics.
