|
VIRGINIJUS ŠIKŠNYS Distinguished Professor |
 |
Phages are the most abundant organisms in the biosphere and the major parasites of bacteria. They infect bacteria in order to replicate and usually kill bacteria when the replication is completed. In response to the phage threat, bacteria developed multiple defence barriers for countering and fighting viral attacks. In the Department of Protein-Nucleic Acids Interactions we aim to understand the structure-function relationships of enzymes and enzyme assemblies that contribute to the bacteria defence systems that target invading nucleic acids. We are particularly interested in the molecular machinery involved in the CRISPR-Cas function and the structural and molecular mechanisms of other antiviral defence systems including prokaryotic Argonautes, BREX, toxin-antitoxin systems and others. We are using X-ray crystallography, mutagenesis and functional biochemical as well as biophysical assays to acquire more information on these systems.
CRISPR-Cas has been recently discovered as a prokaryotic antiviral defence system that hijacks short fragments of invasive DNA as spacers and subsequently uses them as templates to generate specific small RNA molecules that combine with Cas proteins into effector complexes that trigger the degradation of foreign nucleic acid. In this respect, CRISPR-Cas systems constitute an adaptive microbial immune system that provides an acquired resistance against invaders. CRISPR systems are very diverse, and we aim to understand the molecular and structural mechanisms of immunity provided by different CRISPR-Cas systems.
In recent years, we have focused on different aspects of CRISPR-Cas systems, in collaboration with Dr. D. Wigley (Imperial College London), Dr. R. Seidel (Universität Leipzig), Dr. M. D. Szczelkun (Bristol University), Dr. J. Young (Corteva), Dr. C. Venclovas (Vilnius University), Dr. M. Bochtler (IUCMB), Drs. K. Makarova and E. Koonin (NIH). We continue to explore molecular mechanisms behind cyclic oligoadenylate signalling pathway discovered by us in 2017 and other proteins related to CRISPR-Cas or other antiviral defence systems.
SELECTED PUBLICATIONS
- Kazlauskiene, M., Kostiuk, G., Venclovas, Č., Tamulaitis, G., Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017, 357: 605–
- Sasnauskas G, Siksnys V. CRISPR adaptation from a structural perspective. Curr Opin Struct Biol. 2020, 19, 65: 17–
- Makarova, K. S., Timinskas, A., Wolf, Y. I., Gussow, A. B., Siksnys, V., Venclovas, Č., Koonin, E. V. Evolutionary and functional classification of the CARF domain superfamily, key sensors in prokaryotic antivirus defense. Nucleic Acids Res. 2020, 48(16): 8828– doi: 10.1093/nar/gkaa635.
- Songailiene, I., Rutkauskas, M., Sinkunas, T., Manakova, E., Wittig, S., Schmidt, C., Siksnys, V., Seidel, R. Decision-making in cascade complexes harboring crRNAs of altered length. Cell Rep. 2019, 28(12): 3157–e4. doi: 10.1016/j.celrep.2019.08.033.
- Wilkinson, M., Drabavicius, G., Silanskas, A., Gasiunas, G., Siksnys, V., Wigley, D. B. Structure of the DNA-bound spacer capture complex of a type II CRISPR-Cas system. Mol Cell. 2019, 75(1): 90–101.e5. doi: 10.1016/j.molcel.2019.04.020.
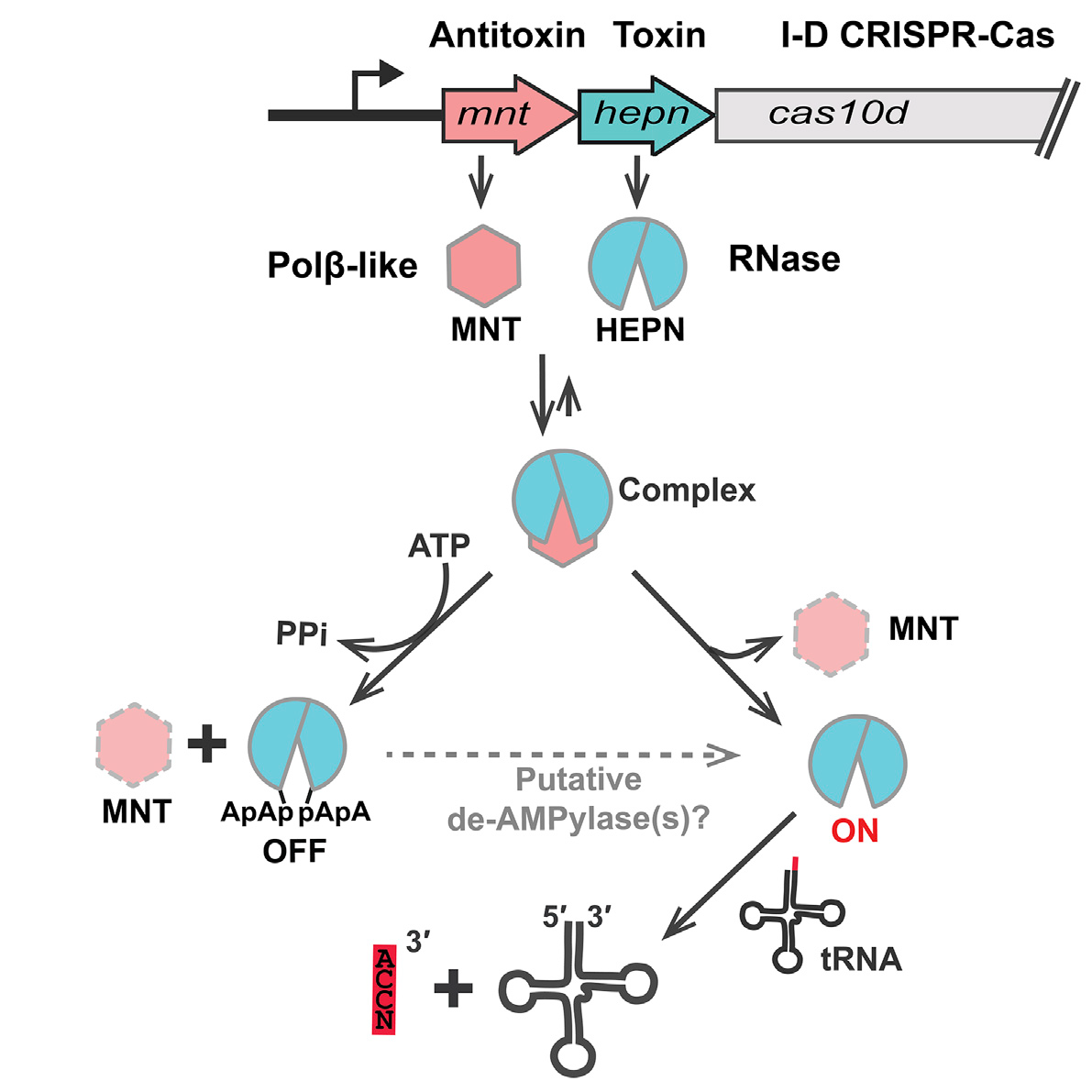
Novel Toxin-Antitoxin System Related to I-D CRISPR-Cas System
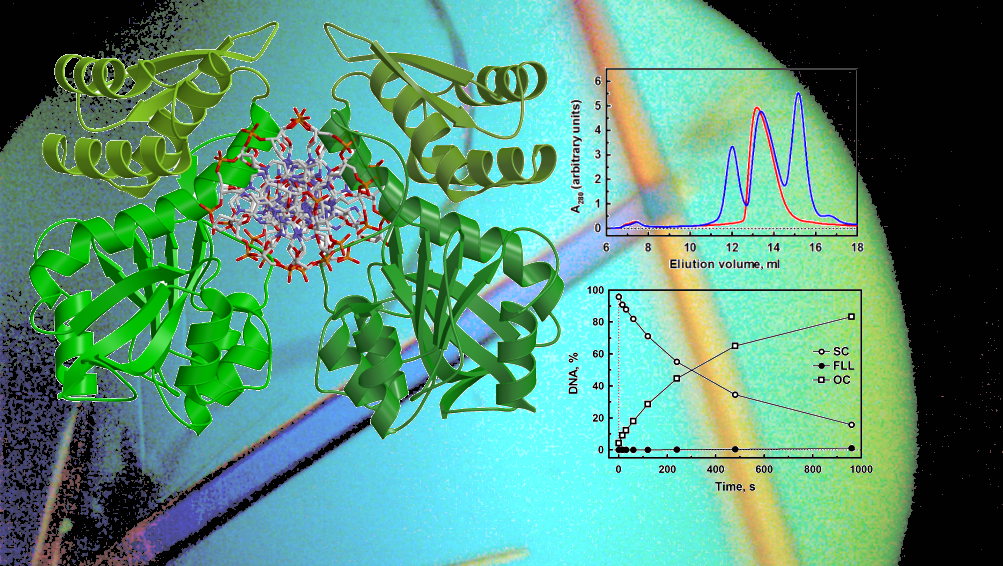
HEPN-MNT toxin-antitoxin (TA) system is encoded in the vicinity of a subtype I-D CRISPR-Cas system in the cyanobacterium Aphanizomenon flos-aquae. Using biochemical and structural methods, we showed that HEPN acts as a toxic RNase, which cleaves off 4 nt from the 3' end in a subset of tRNAs, thereby interfering with translation. Surprisingly, we find that the MNT (minimal nucleotidyltransferase) antitoxin inhibits HEPN RNase through covalent di-AMPylation (diadenylylation) of a conserved tyrosine residue, Y109, in the active site loop. We propose that the HEPN-MNT system functions as a cellular ATP sensor that monitors ATP homeostasis and, at low ATP levels, releases active HEPN toxin. The I-D CRISPR-Cas system present in A. flos-aquae contains a putative Cas3-like ATPase-helicase; thus, the HEPN-MNT TA system could become activated due to additional ATP degradation by CRISPR-Cas3 in response to phage infection. In this case, I-D CRISPR-Cas ATPase-controlled activation of HEPN RNase in A. flos-aquae would be analogous to activation of the auxiliary Csm6 RNase by cyclic oligoadenylate produced by the type III CRISPR-Cas system in response to phage infection. However, the exact mechanism of the HEPN-MNT system action and its possible crosstalk with the CRISPR-Cas system remain to be established (Songailiene et al. Mol. Cell. 2020, S1097-2765(20)30834-0).
|
|
Proposed mechanism of action of A. flos-aquae HEPN-MNT toxin-antitoxin system |
Studies of 5′ Modifications to CRISPR–Cas9 Guide RNA
A key aim in exploiting CRISPR–Cas is guide RNA (gRNA) engineering to introduce additional functionalities, ranging from individual nucleotide changes that increase efficiency of on-target binding to the inclusion of larger functional RNA aptamers or ribonucleoproteins (RNPs). Cas9–gRNA interactions are crucial for complex assembly, but several distinct regions of the gRNA are amenable to modification. We used in vitro ensemble and single-molecule assays to assess the impact of gRNA structural alterations on RNP complex formation, R-loop dynamics, and endonuclease activity. Our results indicate that RNP formation was unaffected by any of our modifications. R-loop formation and DNA cleavage activity were also essentially unaffected by modification of the Upper Stem, first Hairpin and 3′ end. In contrast, we found that 5′ additions of only two or three nucleotides could reduce R-loop formation and cleavage activity of the RuvC domain relative to a single nucleotide addition. Such modifications are a common by-product of in vitro transcribed gRNA. We also observed that addition of a 20 nt RNA hairpin to the 5′ end of a gRNA still supported RNP formation but produced a stable ∼9 bp R-loop that could not activate DNA cleavage. Consideration of these observations will assist in successful gRNA design (Mullally et al., Nucleic Acids Res. 2020, 48, 6811–6823).
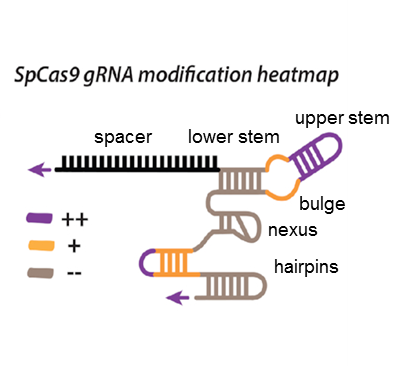
gRNA heatmap showing gRNA regions which tolerate (purple), partially tolerate (orange) and do not tolerate (grey) modification

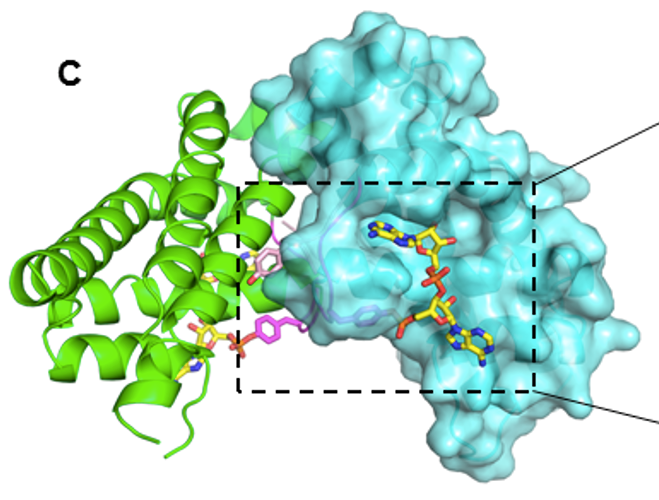 Crystal structure of di_AMPylated HEPN ribonuclease
Crystal structure of di_AMPylated HEPN ribonuclease