|
SAULIUS GRAŽULIS |
|
Crystallography and Molecular Modelling
Modelling matter at atomic level is important for structural biology, material science, physics and (bio)chemistry. These methods become increasingly important with the growth of available computing power, availability of large amounts of high quality, machine-readable computer data and advent of new methods such as machine learning. Our approach to molecular modelling consists of organizing available data into well-defined, curated machine-readable open databases, and then using these databases for scientific inferences applying thoroughly documented, reproducible computation procedures.
The main collection of data that we maintain is the Crystallography Open Database (COD). Over 15 years of development, the COD supervised by the international Advisory Board (of which S. Gražulis and. A. Merkys are members) was transformed into the world’s largest open access small molecule crystal data collection. Containing currently close to half a million records, the COD is widely used by researchers worldwide (the two seminal publications together attracted over 1000 citations), and form basis for extracting scientific knowledge from measurement data. This collection is augmented by well-established databases such as PDB, PubChem, ChEMBL and others.
To perform reproducible computations, our group develops and maintains software tools that are capable of utilizing the Crystallographic Information Framework. These tools are routinely used to ensure the syntactic and semantic validity of data in the COD as well as other projects. Our group also routinely collaborates with the International Union of Crystallography and has contributed to the development of the CIF2 file format and the DDLm dictionary definition language.
Current project is ”Chemical annotation in the Crystallography Open Database (COD)”, 2020–2022 (S-MIP-20-21, project leader - dr. A. Merkys).
SELECTED PUBLICATIONS
1. Vaitkus, A., Merkys, A. & Gražulis, S. Validation of the Crystallography Open Database using the CIF framework. Journal of Applied Crystallography. 2021, accepted.
2. Gražulis, S., Merkys, A., Vaitkus, A., Chateigner, D., Lutterotti, L., Moeck, P., et al., Le Bail, A. Crystallography open database: history, development, and perspectives. In: O. Isayev, A. Tropsha, & S. Curtarolo (Eds.), Materials Informatics. 2019, 1–39. Wiley. doi:10.1002/9783527802265.ch1 .
3. Mendili, Y. E., Vaitkus, A., Merkys, A., Gražulis, S., Chateigner, D., Mathevet, F., et al. Guen, M. L. Raman Open Database: first interconnected Raman–X-ray diffraction open-access resource for material identification. Journal of Applied Crystallography. 2019, 52(3): 618–625. doi:10.1107/s1600576719004229 .
4. Quirós, M., Gražulis, S., Girdzijauskaitė, S., Merkys, A., & Vaitkus, A. Using smiles strings for the description of chemical connectivity in the crystallography open database. Journal of Cheminformatics. 2018, 10(23). doi:10.1186/s13321-018-0279-6.
5. Merkys, A., Mounet, N., Cepellotti, A., Marzari, N., Gražulis, S., Pizzi, G. A posteriori metadata from automated provenance tracking: integration of AiiDA and TCOD. Journal of Cheminformatics. 2017, 9(1): 56. doi:10.1186/s13321-017-0242-y.
Crystallographic Data Validation
The crystallographic data validation topic concerns collection, analysis and validation of crystallographic information in the COD. As experimental crystallographic data is not directly usable in computational chemistry analyses, additional information and assumptions have to be employed to augment the crystallographic data with chemical annotations in fully automated manner. Analysis of the results derived by such processes leads to the identification of outliers, which may be genuine either due to the problems with computation workflows or the data itself. Identification of the latter is crucial to increase the quality of both the crystallographic and the chemical data in the COD as well as other bodies of experimental crystallographic data (project leader - A. Merkys).
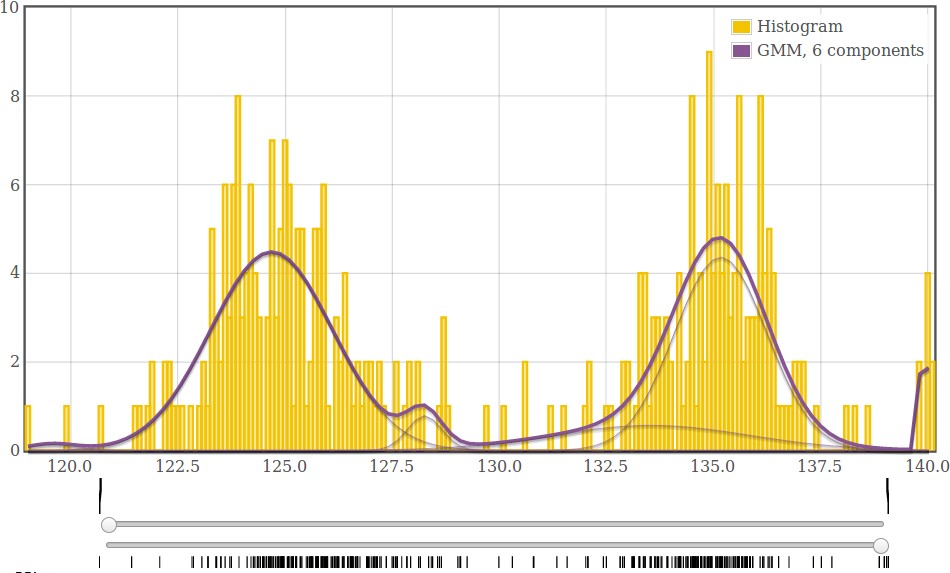
Fig. 1. Distribution of c(cCH)2(H)-c(cCH)2(H)-c(cCH)2(H) bond angles in the COD data.
Derivation of Chemical Information from Crystallographic Data
The emergence of new interdisciplinary fields has stipulated the need to establish a greater connectivity between scientific data from different research areas. One strategy of relating crystallographic data to other fields such as chemistry or material science relies on generating chemical descriptors of molecules from their crystallographic structures and using these descriptors to identify the chemical compounds that the crystals encompass. To facilitate the cross-linking of the COD with other open resources, our group has developed an automated pipeline capable of extrapolating chemical data such as atom connectivity, bond orders and atom charges from crystallographic models, thus enabling the generation of chemical descriptors. These descriptors were later used to link a large portion of the COD to the PubChem database (project leader - A. Vaitkus).
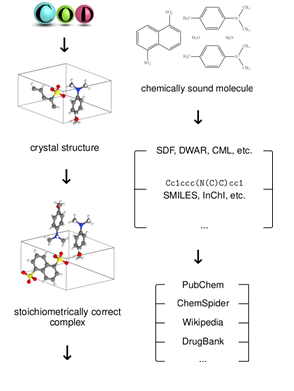
Fig. 2. Schema of the automated pipeline used to derive chemical information from the COD.
Molecular Geometries in Macromolecular Structures
Identifying the probable positions of the protein side-chains is one of the protein modelling steps that can improve the prediction of protein-ligand, protein-protein interactions. In our research, we are trying to approach rotamer library generation problem by scanning for side-chain conformations and calculating potential energy values instead of pooling occurrences of angles only from the structural data (PDB). This enables to study side-chains regardless of unobserved angles or modified amino acids. The flexibility of the method enhances the study of possible side-chain positions and their potential interactions with the ligands (project leader - A. Grybauskas).
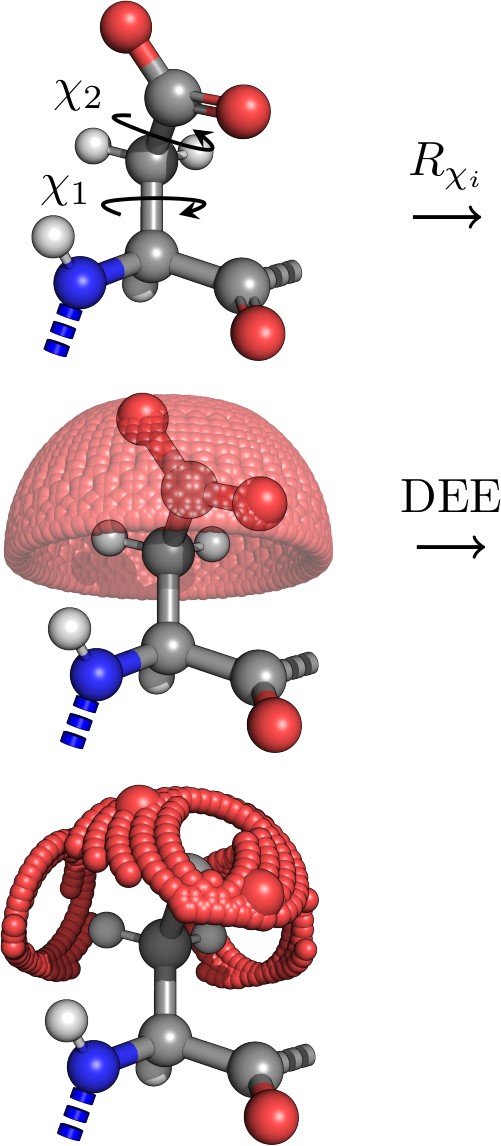
Fig.3. Rotamer generation steps: first, the energy values of all angles are calculated until they reach certain threshold (dead-end elimination), then the lowest energy rotamers are kept.