Structural Biochemistry
Protein X-ray crystallography is the primary technique for elucidation of three-dimensional protein structures, which are critical for understanding macromolecule mechanism and function.
We perform protein crystallization using Oryx8 and Gryphon crystallization robots, monitor crystal growth in automatic Rigaku Minstrel DT UV stations and collect X-ray diffraction data either on an in-house Rigaku MicroMax-007HF X-ray diffractometer fitted with a Dectris Pilatus 3R 200K-A detector or during data collection sessions in DESY synchrotron, Hamburg. A 200 kV Cryo-EM Glacios microscope is currently being installed at the Life Sciences Center.
Our research combines two major directions:
1) Structural characterization of prokaryotic proteins and protein complexes involved in bacterial antiviral defence. In order to survive under a constant pressure of phage infection, bacteria have developed a great variety of defence mechanisms. Currently, we study components of various bacterial antiviral systems, including:
(i) restriction endonucleases (REases), components of Restriction-Modification systems that protect host bacteria by cleaving bacteriophage DNA, constitute a large and highly diverse family of proteins, which differ in their activity regulation, DNA recognition and DNA cleavage mechanisms. We study orthodox Type II enzymes (PfoI [1], Kpn2I, AgeI, BsaWI), ATP-dependent REases (NgoAVII, CglI), and REases specific for methylated DNA sequences (LpnPI, EcoKMcrBC [2], EcoKMcrA [3], the latter in collaboration with Prof. M. Bochtler group from the International Institute of Molecular and Cell Biology in Warsaw);
(ii) components of the CRISPR-Cas adaptive immunity systems (Cas1-Cas2, Cas6, Cascade);
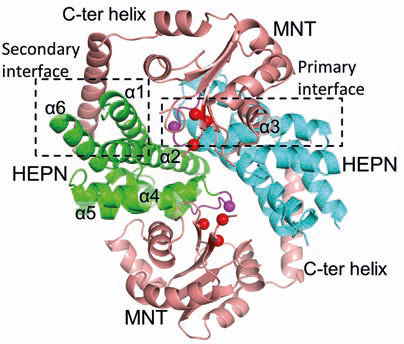
(iii) bacterial toxin-antitoxin systems, e. g. the MNT-HEPN system from A. flos-aquae cyanobacteria [4];
(iv) prokaryotic argonaute proteins and other novel antiviral systems.
2) Crystallographic studies of protein-inhibitor complexes, which are primarily focused on inhibitors of human carbonic anhydrases (hCAs) developed in the Department of Biothermodynamics and Drug Design. hCAs are present in 12 active isoforms in all human tissues. Some isoforms are important therapeutic targets. Design of isoform-specific inhibitors of hCA is a complex project that combines comprehensive thermodynamic description of the protein-ligand interaction with structural characterization. We perform structural characterization of hCA complexes with newly designed inhibitors in order to correlate the binding modes with the thermodynamic parameters of interaction.
SELECTED PUBLICATIONS:
1. Tamulaitiene, G., Manakova, E., Jovaisaite, V., Tamulaitis, G., Grazulis, S., Bochtler, M., Siksnys, V. Unique mechanism of target recognition by PfoI restriction endonuclease of the CCGG-family. Nucleic Acids Research. 2019, 47: 997–1010.
2. Zagorskaitė, E., Manakova, E., Sasnauskas, G. Recognition of modified cytosine variants by the DNA binding domain of methyl-directed endonuclease McrBC. FEBS Letters. 2018, 592: 3335–3345.
3. Slyvka, A., Zagorskaitė, E., Czapinska, H., Sasnauskas, G., Bochtler, M. Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping. Nucleic Acids Research. 2019, 47: 11943–11955.
4. Songailiene, I., Juozapaitis, J., Tamulaitiene, G., Ruksenaite, A., Šulčius, S., Sasnauskas, G., Venclovas, Č., Siksnys, V. HEPN-MNT toxin-antitoxin system: the HEPN ribonuclease is neutralized by oligoAMPylation. Molecular Cell. 2020, 80: 929–1140.
5. Kazokaitė, J., Kairys., V, Smirnovienė, J., Smirnov, A., Manakova, E., Tolvanen, M., Parkkila, S., Matulis, D. Engineered carbonic anhydrase VI-mimic enzyme switched the structure and affinities of inhibitors. Scientific Reports. 2019, 9: 12710.