|
|
RASA PETRAITYTĖ-BURNEIKIENĖ |
RESEARCH OVERVIEW
We investigate aspects related to the production of recombinant proteins in yeast expression systems and the development and optimization of expression systems dedicated to the production of recombinant proteins as virus-like particles (VLPs). VLPs generated in a yeast expression system of viral capsid and envelope proteins have an intrinsic capability of self-assembling into highly organized particles, often without the need for additional viral components. VLPs can induce a strong humoral immune response because of the correct folding of the monomeric proteins, the resulting formation of conformational antigenic determinants and the multimeric structure of identical subunits. Our aim is to understand and compensate the processes in yeast that are triggered by the synthesis of recombinant proteins and to identify the relevant factors for the efficient expression of recombinant proteins. In an attempt to elucidate the requirement of factors for the biosynthesis of recombinant viral and human proteins, we use proteomics, yeast mutant and gene collection studies. Our team is also interested in the search and characterization of new viruses as well as protein engineering based on the construction of chimeric VLPs that harbour foreign epitopes. Yeast-expressed recombinant proteins are applied in the tests for the detection of virus-specific antibodies in human serum and oral fluid samples. A large collection of more than 40 different VLPs derived from various polyomavirus VP1 proteins and papillomavirus 6, 16, 18, 31, 33 L1 proteins were generated. The proteins of measles, mumps, rubella, parainfluenza viruses (1–4), hantaviruses, porcine parvovirus, human bocaviruses (1–4), human metapneumovirus, hepatitis E, and human chaperons (calreticulin and BiP) were produced in yeast cells. Commercially available Microimmune (UK) measles and mumps diagnostic tests are based on the proteins developed in the Department. Moreover, we are focusing on the analysis and research of recombinant biopharmaceutical proteins and recombinant allergen proteins. Our studies include the exploration of a plant expression system for the transient production of a recombinant protein in N. benthamiana. We also concentrate on the research of plant anthocyanin synthesis regulation.
This year Prof. D. Balčiūnas from Temple University, Philadelphia, has joined our group, and new research in the studies of molecular mechanisms of heart regeneration was started.
SELECTED PUBLICATIONS
- Špakova, A., Dalgėdienė, I., Insodaitė, R., Sasnauskienė, A., Žvirblienė, A. and Petraitytė-Burneikienė, R. vBEcoSNBD2 bacteriophage-originated polytubes as a carrier for the presentation of foreign sequences. Virus Research. 2020, 290: 198194. doi: 10.1016/j.virusres.2020.198194.
- Lazutka, J., Simutis, K., Matulis, P., Petraitytė-Burneikienė, R., Kučinskaitė-Kodzė, I., Simanavičius, M. and Tamošiunas, P. L. Antigenicity study of the yeast-generated human parvovirus 4 (PARV4) virus-like particles. Virus Research. 2020, 198236. doi: 10.1016/j.virusres.2020.198236.
- Houen, G., Heiden, J., Trier, N. H., Draborg, A. H., Benros, M. E., Zinkevičiūtė, R., Petraitytė-Burneikienė, R., Ciplys, E., Slibinskas, R. and Frederiksen, J. L. Antibodies to Epstein-Barr virus and neurotropic viruses in multiple sclerosis and optic neuritis. Journal of Neuroimmunology. 2020, 346: 577314. doi: 10.1016/j.jneuroim.2020.577314.
RESEARCH ACTIVITIES in 2020
Molecular Mechanisms of Heart Regeneration. Genetic and Molecular Analysis of the Role of Tbx5a in Heart Regeneration
Cardiovascular disease is one of the leading causes of death in the Western world. Myocardial infarction occurs when blockage in a coronary artery interrupts circulation and blood supply. Oxygen-deprived heart muscle cells die and, if the patient survives, are replaced by a permanent scar, leaving cardiac function impaired. In order to improve regenerative capacity of the human heart, we must first thoroughly understand how this process occurs in an organism, which has innate regenerative ability. We believe that the zebrafish Danio rerio is an excellent model organism to study regeneration. First and foremost, zebrafish possess extensive regenerative capacity, being able to regenerate a variety of injured organs and tissues including the heart. Being a vertebrate, it is much more genetically similar to us humans than a hydra or a planarian. Importantly, a full suite of molecular genetic techniques – some of them developed by us – is available to interrogate gene function in zebrafish.
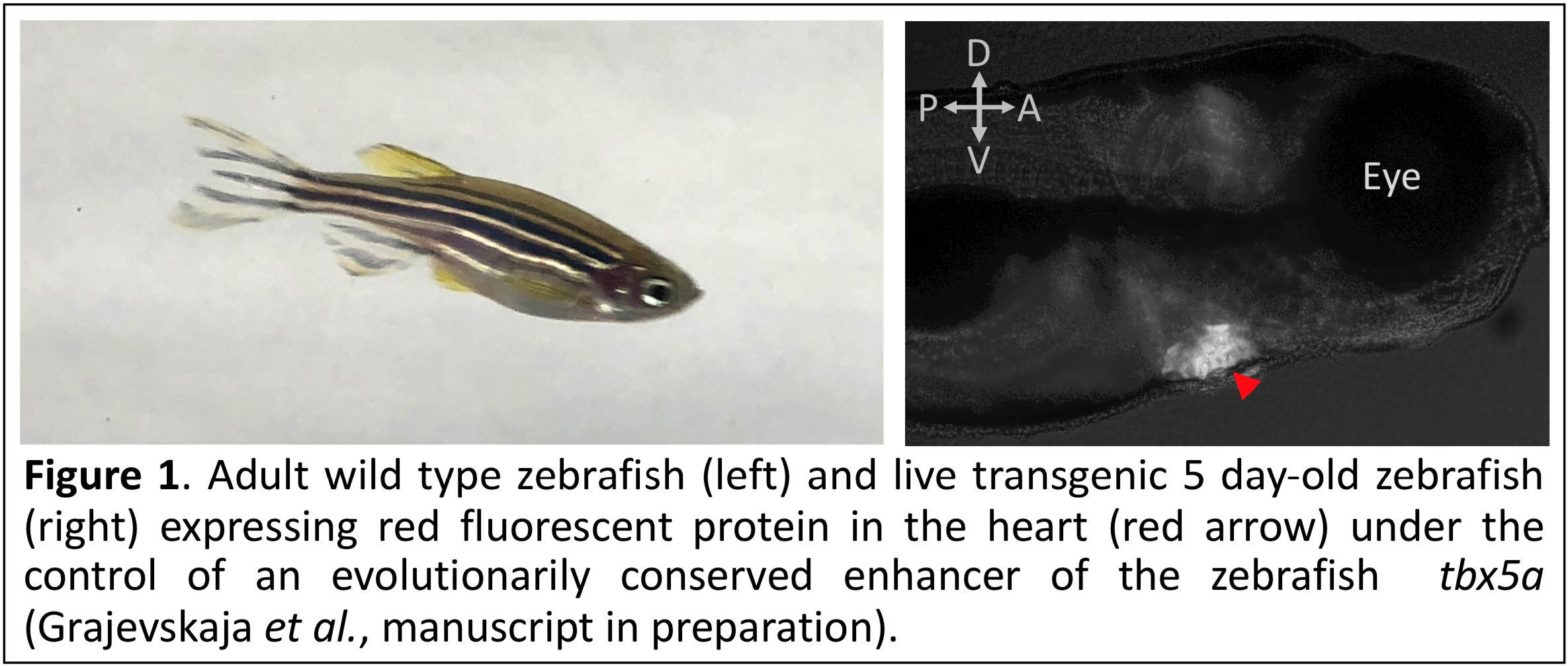
Depending on the type of injury and severity of the injury, regeneration is usually complete in 30–60 days. In our laboratory, we aim to identify transcriptional program required for cardiac regeneration to occur. We are focusing on two highly conserved transcription factors: Tbx5a in the myocardium and Tcf21 in the epicardium. Using a conditional gene trap, we have demonstrated the Tbx5a essential for regeneration to occur (Grajevskaja et al. 2018, PMID: 29933372). In our studies, we are using CRISPR/Cas9 to engineer epitope-tagged alleles (Burg et al. 2016, PMID: 27892520) and conditional mutants (Burg et al. 2018, PMID: 30427827), and characterize these mutants using genomics, developmental biology and molecular biology methods.
New Technologies for Development of Recombinant Allergens
The main goal of this project is to develop a new universal platform for expression and purification of recombinant protein allergens and to prepare open access collection of recombinant allergens including allergen expression plasmids, strains-producers and allergen protein samples. Natural allergens still routinely used are the mixtures of different proteins, but effective allergy diagnosis is requesting precise molecular identification of every analysed allergen protein. The universal platform of the project is based on the fusion of allergen and carrier protein MBP (maltose binding protein). The biosynthesis of recombinant allergens in the most relevant hosts such as bacteria, yeast, plant and mammalian cell cultures are in the process of implementation. The obtained results show that the platform is well applicable for bacterial expression systems (more than 40 allergens tested). Yeast and plant derived recombinant allergens are more sensitive to the host cell-protein modifications different from natural allergens, but still could be applicable in the cases, when bacterial hosts show incorrect alterations in allergen structure. Demonstrating application opportunities of the majority of produced and purified allergens have provided the positive binding results (ELISA and Western blot data) with allergic human IgE antibodies responsible for the allergy diseases. Two new food allergens have been identified from the Lithuanian carp species. Beta-enolase allergen from carp named Cyp c 2 has been included into WHO/IUIS allergen nomenclature data base (http://www.allergen.org/index.php). The conditions for the producing recombinant allergen without MBP (digestion with TEV protease) as well as the conditions for storage and conservation of recombinant allergens have been selected. Constructed allergen expression plasmids have been deposited in the Belgian collection of microorganisms (BCCM; https://bccm.belspo.be/catalogues) for the open-access and public use. Development of advanced molecular technologies of recombinant allergens with high potential for effective allergy diagnosis or immunotherapy will eventually provide superior public and personal health standards.
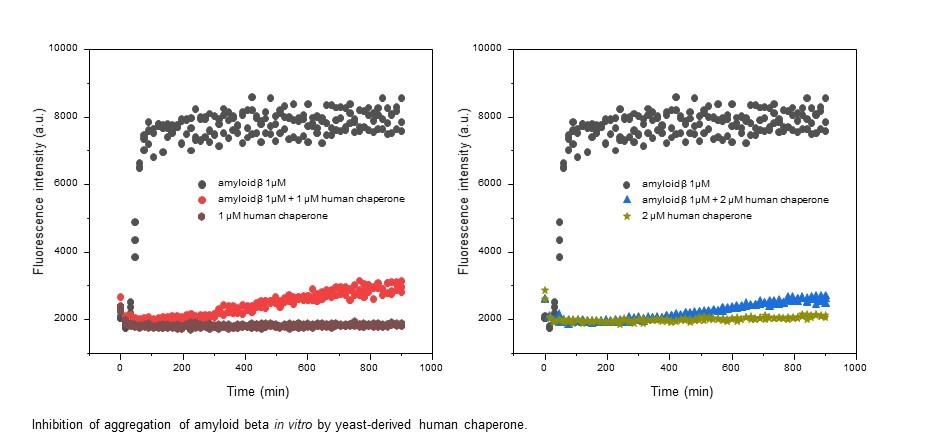
New Methods of Screening for Treatment of Neurodegenerative Disorders
In this project, a range of recombinant human endoplasmic reticulum (ER) chaperones, which potentially could inhibit aggregation of proteins involved in progression of neurodegenerative disorders (ND), are to be tested using both in vitro and in vivo models for these diseases. During the course of the project, we are testing recombinant human ER chaperones in molecular interaction models as potential inhibitors of amyloid-like fibril formation. Inhibition of aggregation will be assessed using several model proteins related to different ND, such as Alzheimer’s and Parkinson’s diseases, amyotrophic lateral sclerosis, prion-related disorders and multiple sclerosis. Chaperones showing the most promising results will be tested alone or in combination with each other in order to evaluate optimal molar ratios of different chaperones. Depending on the results of in vitro studies, specific ND models in experimental animals will be selected for further testing. Moreover, the most efficient ways to deliver recombinant ER chaperones through a blood brain barrier (BBB) and their ability to cross the BBB will be investigated. Furthermore, it will be important to elucidate molecular mechanisms underlying possible positive effects of administered ER chaperones. From many potentially positive results from the in vitro studies, only the most promising options will be selected for the experiments in vivo with the prospect for clinical applications.
Investigation of K. lactis Mutations Conferring Enhanced Secretion Phenotype and Generation of Yeast Strains for Super-secretion of Recombinant Proteins
As the yield of secreted recombinant proteins in yeast is usually far from optimal and needs optimization, the aim of this study was identification and characterization of gene and its mutations conferring the enhanced secretion of phenotype of K. lactis mutant strain MD2/1-9 and application of the acquired knowledge for generation of new yeast super-secretion strains. The mutated gene KlSEC59 encoding dolichol kinase (DK) was identified after sequencing of genomes of both parental and mutated K. lactis strains using the next‐generation sequencing technology. It was confirmed that the double mutant strain with G405S and I419S mutations in DK sequence displayed improved secretion of recombinant proteins despite some glycosylation defects. The introduction of analogical mutations in DK of S. cerevisiae also improved secretion of recombinant proteins but the partial glycosylation deficiency in the double sec 59 mutant strain had stronger impact on yeast cell wall integrity and survival of yeast cells at high temperature compared with the K. lactis Klsec59 mutant. Our results suggested that enhanced secretion related to reduced activity of mutant DK in yeast is most likely related indirectly to lower DK activity and, rather, is the result of mild changes in glycosylation levels and the activity of other proteins in the secretory pathway.
Identification of New Polyomaviruses and Investigation of Their Evolutionary History
Polyomaviruses (PyVs) are widespread, non-enveloped double stranded DNA viruses that infect mammals, birds, and fish but polyomavirus-like sequences have also been recovered from arthropods which points to very ancient association of polyomaviruses with animals. Despite the discovery of numerous PyVs within the last 10 years, still only a very limited number of species have been assessed for the presence and diversity of polyomaviruses (in total, probably fewer than 100), while there are about 5000 mammalian species belonging to 29 orders. Therefore, it is evident that there is a need to identify more PyVs from other host species for a better understanding of overall polyomavirus diversity and evolution, of the long-term dynamics of the coevolution with their hosts or explanation of variations in their toxicity and cross-species transmission.
Analysis of SARS-COV-2 Genomes Isolated from Lithuanian Patients
The aim of this project is sequencing of SARS-COV-2 genomes isolated from Lithuanian patients during the pandemic time and analysing the changes of the virus. The data obtained will be shared by submitting it to public databases such as GISAID. Sequencing of SARS-COV-2 genomes can support ‘genomic epidemiology’- characterizing the virus and helping public health authorities to understand the identity of the virus, whether it is changing and how it is being transmitted - all in conjunction with other epidemiological data. On the other hand, genetic sequencing will help monitor COVID-19 mutations circulating in Lithuania what potentially can allow in time improve diagnostic testing and foresee vaccination effectiveness.

